The Aquilegia genome provides insight into adaptive radiation and reveals an extraordinarily polymorphic chromosome with a unique history
- PMID: 30325307
- PMCID: PMC6255393
- DOI: 10.7554/eLife.36426
The Aquilegia genome provides insight into adaptive radiation and reveals an extraordinarily polymorphic chromosome with a unique history
Abstract
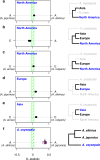
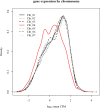
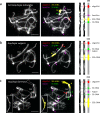
The columbine genus Aquilegia is a classic example of an adaptive radiation, involving a wide variety of pollinators and habitats. Here we present the genome assembly of A. coerulea 'Goldsmith', complemented by high-coverage sequencing data from 10 wild species covering the world-wide distribution. Our analyses reveal extensive allele sharing among species and demonstrate that introgression and selection played a role in the Aquilegia radiation. We also present the remarkable discovery that the evolutionary history of an entire chromosome differs from that of the rest of the genome - a phenomenon that we do not fully understand, but which highlights the need to consider chromosomes in an evolutionary context.
Keywords: Aquilegia; chromosome evolution; chromosomes; gene expression; genetics; genome evolution; genomics; population genetics; speciation.
© 2018, Filiault et al.
Conflict of interest statement
DF, EB, TM, GA, ND, JS, JJ, JG, SS, RH, UH, KB, JY, SM, MK, VN, EK, ML, SH No competing interests declared, MN Reviewing editor, eLife
Figures

















References
-
- Aköz G, Nordborg M. Genome duplication and reorganization in Aquilegia. bioRxiv. 2018 doi: 10.1101/407973~. - DOI
-
- Bastida JM, Alcántara JM, Rey PJ, Vargas P, Herrera CM. Extended phylogeny of Aquilegia: the biogeographical and ecological patterns of two simultaneous but contrasting radiations. Plant Systematics and Evolution. 2010;284:171–185. doi: 10.1007/s00606-009-0243-z. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

