Samp1 Mislocalization in Emery-Dreifuss Muscular Dystrophy
- PMID: 30326651
- PMCID: PMC6210792
- DOI: 10.3390/cells7100170
Samp1 Mislocalization in Emery-Dreifuss Muscular Dystrophy
Abstract
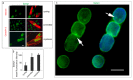
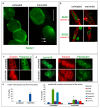
LMNA linked-Emery-Dreifuss muscular dystrophy (EDMD2) is a rare disease characterized by muscle weakness, muscle wasting, and cardiomyopathy with conduction defects. The mutated protein lamin A/C binds several nuclear envelope components including the Linker of Nucleoskeleton and Cytoskeleton (LINC) complex and the inner nuclear membrane protein Samp1 (Spindle Associated Membrane Protein 1). Considering that Samp1 is upregulated during muscle cell differentiation and it is involved in nuclear movement, we hypothesized that it could be part of the protein platform formed by LINC proteins and prelamin A at the myotube nuclear envelope and, as previously demonstrated for those proteins, could be affected in EDMD2. Our results show that Samp1 is uniformly distributed at the nuclear periphery of normal human myotubes and committed myoblasts, but its anchorage at the nuclear poles is related to the presence of farnesylated prelamin A and it is disrupted by the loss of prelamin A farnesylation. Moreover, Samp1 is absent from the nuclear poles in EDMD2 myotubes, which shows that LMNA mutations associated with muscular dystrophy, due to reduced prelamin A levels in muscle cell nuclei, impair Samp1 anchorage. Conversely, SUN1 pathogenetic mutations do not alter Samp1 localization in myotubes, which suggests that Samp1 lies upstream of SUN1 in nuclear envelope protein complexes. The hypothesis that Samp1 is part of the protein platform that regulates microtubule nucleation from the myotube nuclear envelope in concert with pericentrin and LINC components warrants future investigation. As a whole, our data identify Samp1 as a new contributor to EDMD2 pathogenesis and our data are relevant to the understanding of nuclear clustering occurring in laminopathic muscle.
Keywords: Emery-Dreifuss Muscular Dystrophy type 2 (EDMD2); LINC complex; Samp1 (NET5); myonuclear positioning; prelamin A.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

