Electrostatic interactions between middle domain motif-1 and the AAA1 module of the bacterial ClpB chaperone are essential for protein disaggregation
- PMID: 30327424
- PMCID: PMC6302173
- DOI: 10.1074/jbc.RA118.005496
Electrostatic interactions between middle domain motif-1 and the AAA1 module of the bacterial ClpB chaperone are essential for protein disaggregation
Abstract
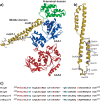
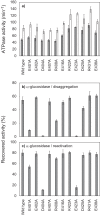
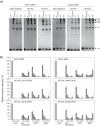
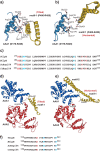
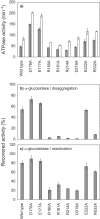
ClpB, a bacterial homologue of heat shock protein 104 (Hsp104), can disentangle aggregated proteins with the help of the DnaK, a bacterial Hsp70, and its co-factors. As a member of the expanded superfamily of ATPases associated with diverse cellular activities (AAA+), ClpB forms a hexameric ring structure, with each protomer containing two AAA+ modules, AAA1 and AAA2. A long coiled-coil middle domain (MD) is present in the C-terminal region of the AAA1 and surrounds the main body of the ring. The MD is subdivided into two oppositely directed short coiled-coils, called motif-1 and motif-2. The MD represses the ATPase activity of ClpB, and this repression is reversed by the binding of DnaK to motif-2. To better understand how the MD regulates ClpB activity, here we investigated the roles of motif-1 in ClpB from Thermus thermophilus (TClpB). Using systematic alanine substitution of the conserved charged residues, we identified functionally important residues in motif-1, and using a photoreactive cross-linker and LC-MS/MS analysis, we further explored potential interacting residues. Moreover, we constructed TClpB mutants in which functionally important residues in motif-1 and in other candidate regions were substituted by oppositely charged residues. These analyses revealed that the intra-subunit pair Glu-401-Arg-532 and the inter-subunit pair Asp-404-Arg-180 are functionally important, electrostatically interacting pairs. Considering these structural findings, we conclude that the Glu-401-Arg-532 interaction shifts the equilibrium of the MD conformation to stabilize the activated form and that the Arg-180-Asp-404 interaction contributes to intersubunit signal transduction, essential for ClpB chaperone activities.
Keywords: ATPases associated with diverse cellular activities (AAA); Hsp104; chaperone; disaggregation; mass spectrometry (MS); protein aggregation; protein conformation; protein cross-linking; protein dynamic; structure-function.
© 2018 Sugita et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures

References
-
- Weibezahn J., Tessarz P., Schlieker C., Zahn R., Maglica Z., Lee S., Zentgraf H., Weber-Ban E. U., Dougan D. A., Tsai F. T., Mogk A., and Bukau B. (2004) Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell 119, 653–665 10.1016/j.cell.2004.11.027 - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

