Characterization of Nigerian breast cancer reveals prevalent homologous recombination deficiency and aggressive molecular features
- PMID: 30327465
- PMCID: PMC6191428
- DOI: 10.1038/s41467-018-06616-0
Characterization of Nigerian breast cancer reveals prevalent homologous recombination deficiency and aggressive molecular features
Erratum in
-
Author Correction: Characterization of Nigerian breast cancer reveals prevalent homologous recombination deficiency and aggressive molecular features.Nat Commun. 2019 Jan 14;10(1):288. doi: 10.1038/s41467-018-07886-4. Nat Commun. 2019. PMID: 30643118 Free PMC article.
Abstract
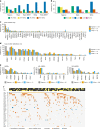
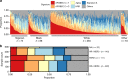
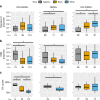
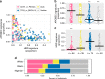
Racial/ethnic disparities in breast cancer mortality continue to widen but genomic studies rarely interrogate breast cancer in diverse populations. Through genome, exome, and RNA sequencing, we examined the molecular features of breast cancers using 194 patients from Nigeria and 1037 patients from The Cancer Genome Atlas (TCGA). Relative to Black and White cohorts in TCGA, Nigerian HR + /HER2 - tumors are characterized by increased homologous recombination deficiency signature, pervasive TP53 mutations, and greater structural variation-indicating aggressive biology. GATA3 mutations are also more frequent in Nigerians regardless of subtype. Higher proportions of APOBEC-mediated substitutions strongly associate with PIK3CA and CDH1 mutations, which are underrepresented in Nigerians and Blacks. PLK2, KDM6A, and B2M are also identified as previously unreported significantly mutated genes in breast cancer. This dataset provides novel insights into potential molecular mechanisms underlying outcome disparities and lay a foundation for deployment of precision therapeutics in underserved populations.
Conflict of interest statement
The authors declare the following competing interests: M.R., A.V., E.L., S.M., R.L., M.M., R.S.J., B.H., KM, M.P.M., W.W., D.P., B.W., and J.B. are all employees of Novartis Institutes for BioMedical Research. K.P.W. serves as President at Tempus. O.I.O is an equity stock holder of CancerIQ and Tempus. All other authors declare no competing interest.
Figures

References
-
- Servick K. Breast cancer. Breast Cancer. 2014;343:1452–1453. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

