Translating genotype data of 44,000 biobank participants into clinical pharmacogenetic recommendations: challenges and solutions
- PMID: 30327539
- PMCID: PMC6752278
- DOI: 10.1038/s41436-018-0337-5
Translating genotype data of 44,000 biobank participants into clinical pharmacogenetic recommendations: challenges and solutions
Abstract
Purpose: Biomedical databases combining electronic medical records and phenotypic and genomic data constitute a powerful resource for the personalization of treatment. To leverage the wealth of information provided, algorithms are required that systematically translate the contained information into treatment recommendations based on existing genotype-phenotype associations.
Methods: We developed and tested algorithms for translation of preexisting genotype data of over 44,000 participants of the Estonian biobank into pharmacogenetic recommendations. We compared the results obtained by genome sequencing, exome sequencing, and genotyping using microarrays, and evaluated the impact of pharmacogenetic reporting based on drug prescription statistics in the Nordic countries and Estonia.
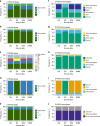
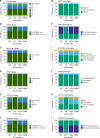
Results: Our most striking result was that the performance of genotyping arrays is similar to that of genome sequencing, whereas exome sequencing is not suitable for pharmacogenetic predictions. Interestingly, 99.8% of all assessed individuals had a genotype associated with increased risks to at least one medication, and thereby the implementation of pharmacogenetic recommendations based on genotyping affects at least 50 daily drug doses per 1000 inhabitants.
Conclusion: We find that microarrays are a cost-effective solution for creating preemptive pharmacogenetic reports, and with slight modifications, existing databases can be applied for automated pharmacogenetic decision support for clinicians.
Keywords: biobank participants; genotyping array; pharmacogenetics; pharmacogenomics; preemptive pharmacogenetic testing.
Conflict of interest statement
V.M.L. is a cofounder and owner of HepaPredict AB. The other authors declare no conflicts of interest.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

