Observation of preQ1-II riboswitch dynamics using single-molecule FRET
- PMID: 30328747
- PMCID: PMC6693549
- DOI: 10.1080/15476286.2018.1536591
Observation of preQ1-II riboswitch dynamics using single-molecule FRET
Abstract
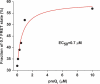
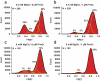
PreQ1 riboswitches regulate the synthesis of the hypermodified tRNA base queuosine by sensing the pyrrolopyrimidine metabolite preQ1. Here, we use single-molecule FRET to interrogate the structural dynamics of apo and preQ1-bound states of the preQ1-II riboswitch from Lactobacillus rhamnosus. We find that the apo-form of the riboswitch spontaneously samples multiple conformations. Magnesium ions and preQ1 stabilize conformations that sequester the ribosome-binding site of the mRNA within the pseudoknotted structure, thus inhibiting translation initiation. Our results reveal that folding of the preQ1-II riboswitch is complex and provide evidence favoring a conformational selection model of effector binding by riboswitches of this class.
Keywords: RNA dynamics; Riboswitch; conformation selection; smFRET.
Figures



References
-
- Belashov IA, Dutta D, Salim M, et al. Tails of three knotty switches: how preQ1 riboswitch structures control protein translation. eLife Sci. 2015. DOI:10.1002/9780470015902.a0021031 - DOI
-
- Sherwood AV, Henkin TM. Riboswitch-mediated gene regulation: novel RNA architectures dictate gene expression responses. Annu Rev Microbiol. 2016;70:361–374. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
