Genetic and epigenetic determinants establish a continuum of Hsf1 occupancy and activity across the yeast genome
- PMID: 30332327
- PMCID: PMC6340206
- DOI: 10.1091/mbc.E18-06-0353
Genetic and epigenetic determinants establish a continuum of Hsf1 occupancy and activity across the yeast genome
Abstract
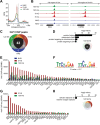
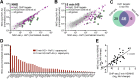
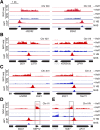
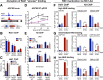
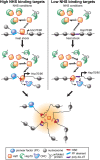
Heat shock factor 1 is the master transcriptional regulator of molecular chaperones and binds to the same cis-acting heat shock element (HSE) across the eukaryotic lineage. In budding yeast, Hsf1 drives the transcription of ∼20 genes essential to maintain proteostasis under basal conditions, yet its specific targets and extent of inducible binding during heat shock remain unclear. Here we combine Hsf1 chromatin immunoprecipitation sequencing (seq), nascent RNA-seq, and Hsf1 nuclear depletion to quantify Hsf1 binding and transcription across the yeast genome. We find that Hsf1 binds 74 loci during acute heat shock, and these are linked to 46 genes with strong Hsf1-dependent expression. Notably, Hsf1's induced DNA binding leads to a disproportionate (∼7.5-fold) increase in nascent transcription. Promoters with high basal Hsf1 occupancy have nucleosome-depleted regions due to the presence of "pioneer factors." These accessible sites are likely critical for Hsf1 occupancy as the activator is incapable of binding HSEs within a stably positioned, reconstituted nucleosome. In response to heat shock, however, Hsf1 accesses nucleosomal sites and promotes chromatin disassembly in concert with the Remodels Structure of Chromatin (RSC) complex. Our data suggest that the interplay between nucleosome positioning, HSE strength, and active Hsf1 levels allows cells to precisely tune expression of the proteostasis network.
Figures


References
-
- Abravaya K, Myers MP, Murphy SP, Morimoto RI. (1992). The human heat shock protein hsp70 interacts with HSF, the transcription factor that regulates heat shock gene expression. Genes Dev , 1153–1164. - PubMed
-
- Cairns BR, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg RD. (1996). RSC, an essential, abundant chromatin-remodeling complex. Cell. , 1249–1260. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

