The Lineage-Defining Transcription Factors SOX2 and NKX2-1 Determine Lung Cancer Cell Fate and Shape the Tumor Immune Microenvironment
- PMID: 30332632
- PMCID: PMC6197489
- DOI: 10.1016/j.immuni.2018.09.020
The Lineage-Defining Transcription Factors SOX2 and NKX2-1 Determine Lung Cancer Cell Fate and Shape the Tumor Immune Microenvironment
Abstract
The major types of non-small-cell lung cancer (NSCLC)-squamous cell carcinoma and adenocarcinoma-have distinct immune microenvironments. We developed a genetic model of squamous NSCLC on the basis of overexpression of the transcription factor Sox2, which specifies lung basal cell fate, and loss of the tumor suppressor Lkb1 (SL mice). SL tumors recapitulated gene-expression and immune-infiltrate features of human squamous NSCLC; such features included enrichment of tumor-associated neutrophils (TANs) and decreased expression of NKX2-1, a transcriptional regulator that specifies alveolar cell fate. In Kras-driven adenocarcinomas, mis-expression of Sox2 or loss of Nkx2-1 led to TAN recruitment. TAN recruitment involved SOX2-mediated production of the chemokine CXCL5. Deletion of Nkx2-1 in SL mice (SNL) revealed that NKX2-1 suppresses SOX2-driven squamous tumorigenesis by repressing adeno-to-squamous transdifferentiation. Depletion of TANs in SNL mice reduced squamous tumors, suggesting that TANs foster squamous cell fate. Thus, lineage-defining transcription factors determine the tumor immune microenvironment, which in turn might impact the nature of the tumor.
Keywords: CXCL5; NKX2-1; SOX2; adenocarcinoma; lung cancer; mouse models; squamous; transdifferentiation; tumor immune microenvironment; tumor-associated neutrophils.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors have no competing interests.
Figures


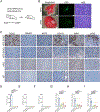


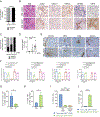

Comment in
-
Lineage Specifiers in Lung Cancer Are Ahead of Their TIME.Immunity. 2018 Oct 16;49(4):587-589. doi: 10.1016/j.immuni.2018.10.004. Immunity. 2018. PMID: 30332625
References
-
- Bezzi M, Seitzer N, Ishikawa T, Reschke M, Chen M, Wang G, Mitchell C, Ng C, Katon J, Lunardi A, et al. (2018). Diverse genetic-driven immune landscapes dictate tumor progression through distinct mechanisms. Nat. Med 24, 165–175. - PubMed
-
- Campbell EL, Bruyninckx WJ, Kelly CJ, Glover LE, McNamee EN, Bowers BE, Bayless AJ, Scully M, Saeedi BJ, Golden-Mason L, et al. (2014). Transmigrating neutrophils shape the mucosal microenvironment through localized oxygen depletion to influence resolution of inflammation. Immunity 40, 66–77. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

