Genetic Architecture of Adaptive Immune System Identifies Key Immune Regulators
- PMID: 30332657
- PMCID: PMC6205839
- DOI: 10.1016/j.celrep.2018.09.048
Genetic Architecture of Adaptive Immune System Identifies Key Immune Regulators
Abstract
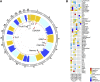
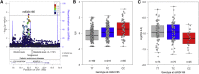
The immune system is highly diverse, but characterization of its genetic architecture has lagged behind the vast progress made by genome-wide association studies (GWASs) of emergent diseases. Our GWAS for 54 functionally relevant phenotypes of the adaptive immune system in 489 healthy individuals identifies eight genome-wide significant associations explaining 6%-20% of variance. Coding and splicing variants in PTPRC and COMMD10 are involved in memory T cell differentiation. Genetic variation controlling disease-relevant T helper cell subsets includes RICTOR and STON2 associated with Th2 and Th17, respectively, and the interferon-lambda locus controlling regulatory T cell proliferation. Early and memory B cell differentiation stages are associated with variation in LARP1B and SP4. Finally, the latrophilin family member ADGRL2 correlates with baseline pro-inflammatory interleukin-6 levels. Suggestive associations reveal mechanisms of autoimmune disease associations, in particular related to pro-inflammatory cytokine production. Pinpointing these key human immune regulators offers attractive therapeutic perspectives.
Keywords: adaptive immune system; association; autoimmunity; genetics; genome-wide association; immune phenotype; susceptibility.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Baurecht H., Hotze M., Brand S., Büning C., Cormican P., Corvin A., Ellinghaus D., Ellinghaus E., Esparza-Gordillo J., Fölster-Holst R., Psoriasis Association Genetics Extension Genome-wide comparative analysis of atopic dermatitis and psoriasis gives insight into opposing genetic mechanisms. Am. J. Hum. Genet. 2015;96:104–120. - PMC - PubMed
-
- Blaeser F., Ho N., Prywes R., Chatila T.A. Ca(2+)-dependent gene expression mediated by MEF2 transcription factors. J. Biol. Chem. 2000;275:197–209. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

