Ciprofloxacin Enhances TRAIL-Induced Apoptosis in Lung Cancer Cells by Upregulating the Expression and Protein Stability of Death Receptors through CHOP Expression
- PMID: 30332761
- PMCID: PMC6214089
- DOI: 10.3390/ijms19103187
Ciprofloxacin Enhances TRAIL-Induced Apoptosis in Lung Cancer Cells by Upregulating the Expression and Protein Stability of Death Receptors through CHOP Expression
Abstract
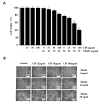
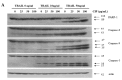
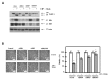
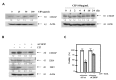
Ciprofloxacin (CIP) is a potent antimicrobial agent with multiple effects on host cells and tissues. Previous studies have highlighted their proapoptotic effect on human cancer cells. The current study showed that subtoxic doses of CIP effectively sensitized multiple cancer cells to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis. Although TRAIL alone mediated the partial proteolytic processing of procaspase-3 in lung cancer cells, co-treatment with CIP and TRAIL efficiently restored the complete activation of caspases. We found that treatment of lung cancer with CIP significantly upregulated the expression and protein stability of death receptor (DR) 5. These effects were mediated through the regulation of transcription factor CCAT enhancer-binding protein homologous protein (CHOP) since the silencing of these signaling molecules abrogated the effect of CIP. Taken together, these results indicated that the upregulation of death receptor expression and protein stability by CIP contributed to the restoration of TRAIL-sensitivity in lung cancer cells.
Keywords: CHOP; TRAIL; ciprofloxacin; death receptor.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Ethanolic extract of Descurainia sophia seeds sensitizes A549 human lung cancer cells to TRAIL cytotoxicity by upregulating death receptors.BMC Complement Altern Med. 2016 Apr 2;16:115. doi: 10.1186/s12906-016-1094-0. BMC Complement Altern Med. 2016. PMID: 27039295 Free PMC article.
-
Arsenic trioxide sensitizes human glioma cells, but not normal astrocytes, to TRAIL-induced apoptosis via CCAAT/enhancer-binding protein homologous protein-dependent DR5 up-regulation.Cancer Res. 2008 Jan 1;68(1):266-75. doi: 10.1158/0008-5472.CAN-07-2444. Cancer Res. 2008. PMID: 18172319
-
Ilimaquinone induces death receptor expression and sensitizes human colon cancer cells to TRAIL-induced apoptosis through activation of ROS-ERK/p38 MAPK-CHOP signaling pathways.Food Chem Toxicol. 2014 Sep;71:51-9. doi: 10.1016/j.fct.2014.06.001. Epub 2014 Jun 12. Food Chem Toxicol. 2014. PMID: 24930757
-
Capsaicin sensitizes malignant glioma cells to TRAIL-mediated apoptosis via DR5 upregulation and survivin downregulation.Carcinogenesis. 2010 Mar;31(3):367-75. doi: 10.1093/carcin/bgp298. Epub 2009 Nov 25. Carcinogenesis. 2010. PMID: 19939880
-
Control of death receptor ligand activity by posttranslational modifications.Cell Mol Life Sci. 2010 May;67(10):1631-42. doi: 10.1007/s00018-010-0289-7. Epub 2010 Mar 20. Cell Mol Life Sci. 2010. PMID: 20306114 Free PMC article. Review.
Cited by
-
Inhalable liposomal powder formulations for co-delivery of synergistic ciprofloxacin and colistin against multi-drug resistant gram-negative lung infections.Int J Pharm. 2020 Feb 15;575:118915. doi: 10.1016/j.ijpharm.2019.118915. Epub 2019 Dec 6. Int J Pharm. 2020. PMID: 31816354 Free PMC article.
-
Dibasic Derivatives of Phenylcarbamic Acid as Prospective Antibacterial Agents Interacting with Cytoplasmic Membrane.Antibiotics (Basel). 2020 Feb 6;9(2):64. doi: 10.3390/antibiotics9020064. Antibiotics (Basel). 2020. PMID: 32041117 Free PMC article.
-
Synthesis, characterization, and toxicity evaluation of ciprofloxacin-chloranilic acid charge transfer complexes: potential for anticancer applications.RSC Med Chem. 2025 Mar 29. doi: 10.1039/d5md00091b. Online ahead of print. RSC Med Chem. 2025. PMID: 40337308 Free PMC article.
-
Upregulation of C/EBP Homologous Protein induced by ER Stress Mediates Epithelial to Myofibroblast Transformation in ADTKD-UMOD.Int J Med Sci. 2022 Jan 24;19(2):364-376. doi: 10.7150/ijms.65036. eCollection 2022. Int J Med Sci. 2022. PMID: 35165522 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

