Genome-wide identification and analysis of the COI gene family in wheat (Triticum aestivum L.)
- PMID: 30332983
- PMCID: PMC6192174
- DOI: 10.1186/s12864-018-5116-9
Genome-wide identification and analysis of the COI gene family in wheat (Triticum aestivum L.)
Abstract
Background: COI (CORONATINE INSENSITIVE), an F-box component of the Skp1-Cullin-F-box protein (SCFCOI1) ubiquitin E3 ligase, plays important roles in the regulation of plant growth and development. Recent studies have shown that COIs are involved in pollen fertility. In this study, we identified and characterized COI genes in the wheat genome and analyzed expression patterns under abiotic stress.
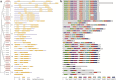
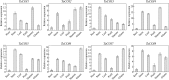
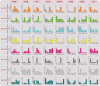
Results: A total of 18 COI candidate sequences for 8 members of COI gene family were isolated in wheat (Triticum aestivum L.). Phylogenetic and structural analyses showed that these COI genes could be divided into seven distinct subfamilies. The COI genes showed high expression in stamens and glumes. The qRT-PCR results revealed that wheat COIs were involved in several abiotic stress responses and anther/glume dehiscence in the photoperiod-temperature sensitive genic male sterile (PTGMS) wheat line BS366.
Conclusions: The structural characteristics and expression patterns of the COI gene family in wheat as well as the stress-responsive and differential tissue-specific expression profiles of each TaCOI gene were examined in PTGMS wheat line BS366. In addition, we examined SA- and MeJA-induced gene expression in the wheat anther and glume to investigate the role of COI in the JA signaling pathway, involved in the regulation of abnormal anther dehiscence in the PTGMS wheat line. The results of this study contribute novel and detailed information about the TaCOI gene family in wheat and could be used as a benchmark for future studies of the molecular mechanisms of PTGMS in other crops.
Keywords: COI; Gene family; JA signaling; Male sterile; Quantitative real-time PCR; Triticum aestivum L..
Conflict of interest statement
Ethics approval and consent to participate
PTGMS wheat line BS366 seeds were provided by Beijing Engineering and Technique Research Center for Hybrid Wheat.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures






References
-
- Sasaki Y, Asamizu E, Shibata D, Nakamura Y, Kaneko T, Awai K, Amagai M, Kuwata C, Tsugane T, Masuda T. Monitoring of methyl jasmonate-responsive genes in Arabidopsis by cDNA macroarray: self-activation of jasmonic acid biosynthesis and crosstalk with other phytohormone signaling pathways. DNA Res. 2001;8(4):153. doi: 10.1093/dnares/8.4.153. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

