Methods used in the spatial analysis of tuberculosis epidemiology: a systematic review
- PMID: 30333043
- PMCID: PMC6193308
- DOI: 10.1186/s12916-018-1178-4
Methods used in the spatial analysis of tuberculosis epidemiology: a systematic review
Abstract
Background: Tuberculosis (TB) transmission often occurs within a household or community, leading to heterogeneous spatial patterns. However, apparent spatial clustering of TB could reflect ongoing transmission or co-location of risk factors and can vary considerably depending on the type of data available, the analysis methods employed and the dynamics of the underlying population. Thus, we aimed to review methodological approaches used in the spatial analysis of TB burden.
Methods: We conducted a systematic literature search of spatial studies of TB published in English using Medline, Embase, PsycInfo, Scopus and Web of Science databases with no date restriction from inception to 15 February 2017. The protocol for this systematic review was prospectively registered with PROSPERO ( CRD42016036655 ).
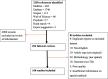
Results: We identified 168 eligible studies with spatial methods used to describe the spatial distribution (n = 154), spatial clusters (n = 73), predictors of spatial patterns (n = 64), the role of congregate settings (n = 3) and the household (n = 2) on TB transmission. Molecular techniques combined with geospatial methods were used by 25 studies to compare the role of transmission to reactivation as a driver of TB spatial distribution, finding that geospatial hotspots are not necessarily areas of recent transmission. Almost all studies used notification data for spatial analysis (161 of 168), although none accounted for undetected cases. The most common data visualisation technique was notification rate mapping, and the use of smoothing techniques was uncommon. Spatial clusters were identified using a range of methods, with the most commonly employed being Kulldorff's spatial scan statistic followed by local Moran's I and Getis and Ord's local Gi(d) tests. In the 11 papers that compared two such methods using a single dataset, the clustering patterns identified were often inconsistent. Classical regression models that did not account for spatial dependence were commonly used to predict spatial TB risk. In all included studies, TB showed a heterogeneous spatial pattern at each geographic resolution level examined.
Conclusions: A range of spatial analysis methodologies has been employed in divergent contexts, with all studies demonstrating significant heterogeneity in spatial TB distribution. Future studies are needed to define the optimal method for each context and should account for unreported cases when using notification data where possible. Future studies combining genotypic and geospatial techniques with epidemiologically linked cases have the potential to provide further insights and improve TB control.
Keywords: Genotypic cluster; Spatial analysis; Tuberculosis.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
-
- Haase I, Olson S, Behr MA, Wanyeki I, Thibert L, Scott A, Zwerling A, Ross N, Brassard P, Menzies D, et al. Use of geographic and genotyping tools to characterise tuberculosis transmission in Montreal. Int J Tuber Lung Dis. 2007;11(6):632–638. - PubMed
-
- Dye C, Loyd K. Tuberculosis. In: Jamison DTBJ, Measham AR, editors. Disease control priorities in developing countries. 2. Washington DC: WorldBank; 2006. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical