Multiple Introductions and Antigenic Mismatch with Vaccines May Contribute to Increased Predominance of G12P[8] Rotaviruses in the United States
- PMID: 30333170
- PMCID: PMC6288334
- DOI: 10.1128/JVI.01476-18
Multiple Introductions and Antigenic Mismatch with Vaccines May Contribute to Increased Predominance of G12P[8] Rotaviruses in the United States
Abstract
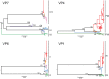
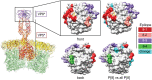
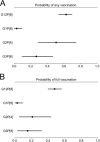
Rotavirus is the leading global cause of diarrheal mortality for unvaccinated children under 5 years of age. The outer capsid of rotavirus virions consists of VP7 and VP4 proteins, which determine viral G and P types, respectively, and are primary targets of neutralizing antibodies. Successful vaccination depends upon generating broadly protective immune responses following exposure to rotaviruses presenting a limited number of G- and P-type antigens. Vaccine introduction resulted in decreased rotavirus disease burden but also coincided with the emergence of uncommon G and P genotypes, including G12. To gain insight into the recent predominance of G12P[8] rotaviruses in the United States, we evaluated 142 complete rotavirus genome sequences and metadata from 151 clinical specimens collected in Nashville, TN, from 2011 to 2013 through the New Vaccine Surveillance Network. Circulating G12P[8] strains were found to share many segments with other locally circulating strains but to have distinct constellations. Phylogenetic analyses of G12 sequences and their geographic sources provided evidence for multiple separate introductions of G12 segments into Nashville, TN. Antigenic epitopes of VP7 proteins of G12P[8] strains circulating in Nashville, TN, differ markedly from those of vaccine strains. Fully vaccinated children were found to be infected with G12P[8] strains more frequently than with other rotavirus genotypes. Multiple introductions and significant antigenic mismatch may in part explain the recent predominance of G12P[8] strains in the United States and emphasize the need for continued monitoring of rotavirus vaccine efficacy against emerging rotavirus genotypes.IMPORTANCE Rotavirus is an important cause of childhood diarrheal disease worldwide. Two immunodominant proteins of rotavirus, VP7 and VP4, determine G and P genotypes, respectively. Recently, G12P[8] rotaviruses have become increasingly predominant. By analyzing rotavirus genome sequences from stool specimens obtained in Nashville, TN, from 2011 to 2013 and globally circulating rotaviruses, we found evidence of multiple introductions of G12 genes into the area. Based on sequence polymorphisms, VP7 proteins of these viruses are predicted to present themselves to the immune system very differently than those of vaccine strains. Many of the sick children with G12P[8] rotavirus in their diarrheal stools also were fully vaccinated. Our findings emphasize the need for continued monitoring of circulating rotaviruses and the effectiveness of the vaccines against strains with emerging G and P genotypes.
Keywords: G12P[8]; efficacy; genotype; phylogenetics; rotavirus; vaccine.
Copyright © 2018 American Society for Microbiology.
Figures



Similar articles
-
Unexpected spreading of G12P[8] rotavirus strains among young children in a small area of central Italy.J Med Virol. 2015 Aug;87(8):1292-302. doi: 10.1002/jmv.24180. Epub 2015 Mar 10. J Med Virol. 2015. PMID: 25758365
-
Genetic analyses reveal differences in the VP7 and VP4 antigenic epitopes between human rotaviruses circulating in Belgium and rotaviruses in Rotarix and RotaTeq.J Clin Microbiol. 2012 Mar;50(3):966-76. doi: 10.1128/JCM.05590-11. Epub 2011 Dec 21. J Clin Microbiol. 2012. PMID: 22189107 Free PMC article.
-
Phylogenetic analysis of VP7 and VP4 genes of the most predominant human group A rotavirus G12 identified in children with acute gastroenteritis in Himachal Pradesh, India during 2013-2016.J Med Virol. 2021 Nov;93(11):6200-6209. doi: 10.1002/jmv.27142. Epub 2021 Jun 24. J Med Virol. 2021. PMID: 34138482
-
Genetic diversity of G1P[8] rotavirus VP7 and VP8* antigens in Finland over a 20-year period: No evidence for selection pressure by universal mass vaccination with RotaTeq® vaccine.Infect Genet Evol. 2013 Oct;19:51-8. doi: 10.1016/j.meegid.2013.06.026. Epub 2013 Jul 4. Infect Genet Evol. 2013. PMID: 23831933 Review.
-
Circulating rotavirus strains in children with acute gastroenteritis in Iran, 1986 to 2023 and their genetic/antigenic divergence compared to approved vaccines strains (Rotarix, RotaTeq, ROTAVAC, ROTASIIL) before mass vaccination: Clues for vaccination policy makers.Virus Res. 2024 Aug;346:199411. doi: 10.1016/j.virusres.2024.199411. Epub 2024 Jun 3. Virus Res. 2024. PMID: 38823689 Free PMC article. Review.
Cited by
-
Product review of the rotavirus vaccines ROTASIIL, ROTAVAC, and Rotavin-M1.Hum Vaccin Immunother. 2021 Apr 3;17(4):1223-1234. doi: 10.1080/21645515.2020.1804245. Epub 2020 Oct 29. Hum Vaccin Immunother. 2021. PMID: 33121329 Free PMC article. Review.
-
Driving forces of continuing evolution of rotaviruses.World J Virol. 2024 Jun 25;13(2):93774. doi: 10.5501/wjv.v13.i2.93774. World J Virol. 2024. PMID: 38984077 Free PMC article. Review.
-
Group A rotavirus surveillance before vaccine introduction in Italy, September 2014 to August 2017.Euro Surveill. 2019 Apr;24(15):1800418. doi: 10.2807/1560-7917.ES.2019.24.15.1800418. Euro Surveill. 2019. PMID: 30994104 Free PMC article.
-
Exploring rotavirus proteome to identify potential B- and T-cell epitope using computational immunoinformatics.Heliyon. 2020 Dec 29;6(12):e05760. doi: 10.1016/j.heliyon.2020.e05760. eCollection 2020 Dec. Heliyon. 2020. PMID: 33426322 Free PMC article.
-
Human Rotaviruses of Multiple Genotypes Acquire Conserved VP4 Mutations during Serial Passage.Viruses. 2024 Jun 18;16(6):978. doi: 10.3390/v16060978. Viruses. 2024. PMID: 38932271 Free PMC article.
References
-
- Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, McGee L, von Gottberg A, Song JH, Ko KS, Pichon B, Baker S, Parry CM, Lambertsen LM, Shahinas D, Pillai DR, Mitchell TJ, Dougan G, Tomasz A, Klugman KP, Parkhill J, Hanage WP, Bentley SD. 2011. Rapid pneumococcal evolution in response to clinical interventions. Science 331:430–434. doi:10.1126/science.1198545. - DOI - PMC - PubMed
-
- Lam C, Octavia S, Ricafort L, Sintchenko V, Gilbert GL, Wood N, McIntyre P, Marshall H, Guiso N, Keil AD, Lawrence A, Robson J, Hogg G, Lan R. 2014. Rapid increase in pertactin-deficient Bordetella pertussis isolates, Australia. Emerg Infect Dis 20:626–633. doi:10.3201/eid2004.131478. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

