The C Terminus of Rotavirus VP4 Protein Contains an Actin Binding Domain Which Requires Cooperation with the Coiled-Coil Domain for Actin Remodeling
- PMID: 30333172
- PMCID: PMC6288328
- DOI: 10.1128/JVI.01598-18
The C Terminus of Rotavirus VP4 Protein Contains an Actin Binding Domain Which Requires Cooperation with the Coiled-Coil Domain for Actin Remodeling
Abstract
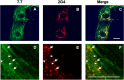
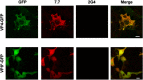
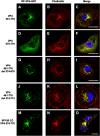
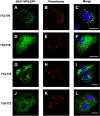
The interactions between viruses and actin cytoskeleton have been widely studied. We showed that rotaviruses remodel microfilaments in intestinal cells and demonstrated that this was due to the VP4 spike protein. Microfilaments mainly occur in the apical domain of infected polarized enterocytes and favor the polarized apical exit of viral progeny. The present work aims at the identification of molecular determinants of actin-VP4 interactions. We used various deletion mutants of VP4 that were transfected into Cos-7 cells and analyzed interactions by immunofluorescence confocal microscopy. It has been established that the C-terminal part of VP4 is embedded within viral particles when rotavirus assembles. The use of specific monoclonal antibodies demonstrated that VP4 is expressed in different forms in infected cells: classically as spike on the outer layer of virus particles, but also as free soluble protein in the cytosol. The C terminus of free VP4 was identified as interacting with actin microfilaments. The VP4 actin binding domain is unable to promote microfilament remodeling by itself; the coiled-coil domain is also required in this process. This actin-binding domain was shown to dominate a previously identified peroxisomal targeting signal, located in the three last amino acids of VP4. The newly identified actin-binding domain is highly conserved in rotavirus strains from species A, B, and C, suggesting that actin binding and remodeling is a general strategy for rotavirus exit. This provides a novel mechanism of protein-protein interactions, not involving cell signaling pathways, to facilitate rotavirus exit.IMPORTANCE Rotaviruses are causal agents of acute infantile viral diarrhea. In intestinal cells, in vitro as well as in vivo, virus assembly and exit do not imply cell lysis but rely on an active process in which the cytoskeleton plays a major role. We describe here a novel molecular mechanism by which the rotavirus spike protein VP4 drives actin remodeling. This relies on the fact that VP4 occurs in different forms. Besides its structural function within the virion, a large proportion of VP4 is expressed as free protein. Here, we show that free VP4 possesses a functional actin-binding domain. This domain, in coordination with a coiled-coil domain, promotes actin cytoskeleton remodeling, thereby providing the capacity to destabilize the cell membrane and allow efficient rotavirus exit.
Keywords: VP4 protein; actin binding; actin remodeling; protein domain; rotavirus.
Copyright © 2018 American Society for Microbiology.
Figures





Similar articles
-
Role for actin in the polarized release of rotavirus.J Virol. 2007 May;81(9):4892-4. doi: 10.1128/JVI.02698-06. Epub 2007 Feb 14. J Virol. 2007. PMID: 17301135 Free PMC article.
-
Actin-Dependent Nonlytic Rotavirus Exit and Infectious Virus Morphogenetic Pathway in Nonpolarized Cells.J Virol. 2018 Feb 26;92(6):e02076-17. doi: 10.1128/JVI.02076-17. Print 2018 Mar 15. J Virol. 2018. PMID: 29263265 Free PMC article.
-
Rotavirus spike protein VP4 binds to and remodels actin bundles of the epithelial brush border into actin bodies.J Virol. 2006 Apr;80(8):3947-56. doi: 10.1128/JVI.80.8.3947-3956.2006. J Virol. 2006. PMID: 16571811 Free PMC article.
-
Rotavirus assembly: an alternative model that utilizes an atypical trafficking pathway.Curr Top Microbiol Immunol. 2006;309:245-61. doi: 10.1007/3-540-30773-7_9. Curr Top Microbiol Immunol. 2006. PMID: 16909902 Free PMC article. Review.
-
Get in and get out: Remodeling of the cellular actin cytoskeleton upon HIV-1 infection.Biol Cell. 2023 Apr;115(4):e2200085. doi: 10.1111/boc.202200085. Epub 2023 Feb 15. Biol Cell. 2023. PMID: 36597754 Review.
Cited by
-
Advances in the Development of Antiviral Compounds for Rotavirus Infections.mBio. 2021 May 11;12(3):e00111-21. doi: 10.1128/mBio.00111-21. mBio. 2021. PMID: 33975930 Free PMC article. Review.
-
The Eukaryotic Translation Initiation Factor 4F Complex Restricts Rotavirus Infection via Regulating the Expression of IRF1 and IRF7.Int J Mol Sci. 2019 Mar 29;20(7):1580. doi: 10.3390/ijms20071580. Int J Mol Sci. 2019. PMID: 30934842 Free PMC article.
-
Rotavirus Spike Protein VP4 Mediates Viroplasm Assembly by Association to Actin Filaments.J Virol. 2022 Sep 14;96(17):e0107422. doi: 10.1128/jvi.01074-22. Epub 2022 Aug 8. J Virol. 2022. PMID: 35938869 Free PMC article.
-
Identification and characterization of coiled-coil motifs across Autographa californica multiple nucleopolyhedrovirus genome.Heliyon. 2022 Sep 10;8(9):e10588. doi: 10.1016/j.heliyon.2022.e10588. eCollection 2022 Sep. Heliyon. 2022. PMID: 36132175 Free PMC article.
-
The Role of the Host Cytoskeleton in the Formation and Dynamics of Rotavirus Viroplasms.Viruses. 2024 Apr 25;16(5):668. doi: 10.3390/v16050668. Viruses. 2024. PMID: 38793550 Free PMC article. Review.
References
-
- Pesavento JB, Crawford SE, Estes MK, Prasad BVV. 2006. Rotavirus proteins: structure and assembly. Curr Top Microbiol Immunol 309:189–219. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

