Polyamines in protozoan pathogens
- PMID: 30333232
- PMCID: PMC6290135
- DOI: 10.1074/jbc.TM118.003342
Polyamines in protozoan pathogens
Abstract
Polyamines are polycationic organic amines that are required for all eukaryotic life, exemplified by the polyamine spermidine, which plays an essential role in translation. They also play more specialized roles that differ across species, and their chemical versatility has been fully exploited during the evolution of protozoan pathogens. These eukaryotic pathogens, which cause some of the most globally widespread infectious diseases, have acquired species-specific polyamine-derived metabolites with essential cellular functions and have evolved unique mechanisms that regulate their core polyamine biosynthetic pathways. Many of these parasitic species have lost enzymes and or transporters from the polyamine metabolic pathway that are found in the human host. These pathway differences have prompted drug discovery efforts to target the parasite polyamine pathways, and indeed, the only clinically approved drug targeting the polyamine biosynthetic pathway is used to manage human African trypanosomiasis. This Minireview will primarily focus on polyamine metabolism and function in Trypanosoma, Leishmania, and Plasmodium species, which are the causative agents of human African trypanosomiasis (HAT) and Chagas disease, Leishmaniasis, and malaria, respectively. Aspects of polyamine metabolism across a diverse group of protozoan pathogens will also be explored.
Keywords: eukaryotic initiation factor 5A (eIF5A); plasmodium; polyamine; protozoan; pseudoenzyme; spermidine; trypanosome.
© 2018 Phillips.
Conflict of interest statement
The author declares that she has no conflicts of interest with the contents of this article
Figures
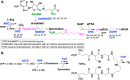

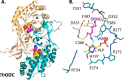
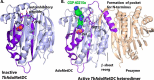

References
-
- Li B., Kim S. H., Zhang Y., Hanfrey C. C., Elliott K. A., Ealick S. E., and Michael A. J. (2015) Different polyamine pathways from bacteria have replaced eukaryotic spermidine biosynthesis in ciliates Tetrahymena thermophila and Paramecium tetaurelia. Mol. Microbiol. 97, 791–807 10.1111/mmi.13066 - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

