Mixed-effects association of single cells identifies an expanded effector CD4+ T cell subset in rheumatoid arthritis
- PMID: 30333237
- PMCID: PMC6448773
- DOI: 10.1126/scitranslmed.aaq0305
Mixed-effects association of single cells identifies an expanded effector CD4+ T cell subset in rheumatoid arthritis
Abstract
High-dimensional single-cell analyses have improved the ability to resolve complex mixtures of cells from human disease samples; however, identifying disease-associated cell types or cell states in patient samples remains challenging because of technical and interindividual variation. Here, we present mixed-effects modeling of associations of single cells (MASC), a reverse single-cell association strategy for testing whether case-control status influences the membership of single cells in any of multiple cellular subsets while accounting for technical confounders and biological variation. Applying MASC to mass cytometry analyses of CD4+ T cells from the blood of rheumatoid arthritis (RA) patients and controls revealed a significantly expanded population of CD4+ T cells, identified as CD27- HLA-DR+ effector memory cells, in RA patients (odds ratio, 1.7; P = 1.1 × 10-3). The frequency of CD27- HLA-DR+ cells was similarly elevated in blood samples from a second RA patient cohort, and CD27- HLA-DR+ cell frequency decreased in RA patients who responded to immunosuppressive therapy. Mass cytometry and flow cytometry analyses indicated that CD27- HLA-DR+ cells were associated with RA (meta-analysis P = 2.3 × 10-4). Compared to peripheral blood, synovial fluid and synovial tissue samples from RA patients contained about fivefold higher frequencies of CD27- HLA-DR+ cells, which comprised ~10% of synovial CD4+ T cells. CD27- HLA-DR+ cells expressed a distinctive effector memory transcriptomic program with T helper 1 (TH1)- and cytotoxicity-associated features and produced abundant interferon-γ (IFN-γ) and granzyme A protein upon stimulation. We propose that MASC is a broadly applicable method to identify disease-associated cell populations in high-dimensional single-cell data.
Copyright © 2018 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Competing interests
All authors declare that they have no competing financial interests. I.K. has been a paid bioinformatics consultant for Outlier Bio LLC since November 2017.
Figures
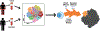



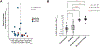

References
-
- Cross M, Smith E, Hoy D, Carmona L, Wolfe F, Vos T, Williams B, Gabriel S, Lassere M, Johns N, Buchbinder R, Woolf A, March L, The global burden of rheumatoid arthritis: estimates from the global burden of disease 2010 study, Annals of the Rheumatic Diseases 73, 1316–1322 (2014). - PubMed
-
- Gizinski AM, Fox DA, T cell subsets and their role in the pathogenesis of rheumatic disease, Current Opinion in Rheumatology 26, 204–210 (2014). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

