Functional genomic landscape of acute myeloid leukaemia
- PMID: 30333627
- PMCID: PMC6280667
- DOI: 10.1038/s41586-018-0623-z
Functional genomic landscape of acute myeloid leukaemia
Abstract
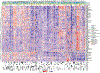
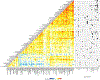
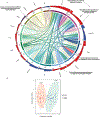
The implementation of targeted therapies for acute myeloid leukaemia (AML) has been challenging because of the complex mutational patterns within and across patients as well as a dearth of pharmacologic agents for most mutational events. Here we report initial findings from the Beat AML programme on a cohort of 672 tumour specimens collected from 562 patients. We assessed these specimens using whole-exome sequencing, RNA sequencing and analyses of ex vivo drug sensitivity. Our data reveal mutational events that have not previously been detected in AML. We show that the response to drugs is associated with mutational status, including instances of drug sensitivity that are specific to combinatorial mutational events. Integration with RNA sequencing also revealed gene expression signatures, which predict a role for specific gene networks in the drug response. Collectively, we have generated a dataset-accessible through the Beat AML data viewer (Vizome)-that can be leveraged to address clinical, genomic, transcriptomic and functional analyses of the biology of AML.
Figures








Comment in
-
Data Offer New Insights into AML.Cancer Discov. 2018 Dec;8(12):1498-1499. doi: 10.1158/2159-8290.CD-NB2018-147. Epub 2018 Nov 6. Cancer Discov. 2018. PMID: 30401682
References
-
- Jemal A, Siegel R, Xu J & Ward E Cancer statistics, 2010. CA: a cancer journal for clinicians 60, 277–300, (2010). - PubMed
-
- National Cancer Institute: Surveillance, Epidemiology, and End Results Program, <https://seer.cancer.gov/statfacts/html/amyl.html> (2018).
-
- Arber DA et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 127, 2391–2405, (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- KL2 TR002370/TR/NCATS NIH HHS/United States
- UL1 TR002369/TR/NCATS NIH HHS/United States
- U54 CA224019/CA/NCI NIH HHS/United States
- R01 CA214428/CA/NCI NIH HHS/United States
- KL2 TR000152/TR/NCATS NIH HHS/United States
- R01 CA183974/CA/NCI NIH HHS/United States
- R37 AI110985/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- T15 LM007088/LM/NLM NIH HHS/United States
- U01 CA217862/CA/NCI NIH HHS/United States
- K99 CA237630/CA/NCI NIH HHS/United States
- UL1 TR002366/TR/NCATS NIH HHS/United States
- P30 CA069533/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

