Tracing Antibody Repertoire Evolution by Systems Phylogeny
- PMID: 30333820
- PMCID: PMC6176079
- DOI: 10.3389/fimmu.2018.02149
Tracing Antibody Repertoire Evolution by Systems Phylogeny
Abstract
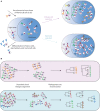
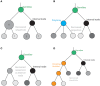
Antibody evolution studies have been traditionally limited to either tracing a single clonal lineage (B cells derived from a single V-(D)-J recombination) over time or examining bulk functionality changes (e.g., tracing serum polyclonal antibody proteins). Studying a single B cell disregards the majority of the humoral immune response, whereas bulk functional studies lack the necessary resolution to analyze the co-existing clonal diversity. Recent advances in high-throughput sequencing (HTS) technologies and bioinformatics have made it possible to examine multiple co-evolving antibody monoclonal lineages within the context of a single repertoire. A plethora of accompanying methods and tools have been introduced in hopes of better understanding how pathogen presence dictates the global evolution of the antibody repertoire. Here, we provide a comprehensive summary of the tremendous progress of this newly emerging field of systems phylogeny of antibody responses. We present an overview encompassing the historical developments of repertoire phylogenetics, state-of-the-art tools, and an outlook on the future directions of this fast-advancing and promising field.
Keywords: B cell evolution; Ig-Seq; antibody lineage; phylogenetics; systems immunology.
Figures
Similar articles
-
Combining mutation and recombination statistics to infer clonal families in antibody repertoires.Elife. 2024 Aug 9;13:e86181. doi: 10.7554/eLife.86181. Elife. 2024. PMID: 39120133 Free PMC article.
-
Comparison of methods for phylogenetic B-cell lineage inference using time-resolved antibody repertoire simulations (AbSim).Bioinformatics. 2017 Dec 15;33(24):3938-3946. doi: 10.1093/bioinformatics/btx533. Bioinformatics. 2017. PMID: 28968873
-
On being the right size: antibody repertoire formation in the mouse and human.Immunogenetics. 2018 Mar;70(3):143-158. doi: 10.1007/s00251-017-1049-8. Epub 2017 Dec 19. Immunogenetics. 2018. PMID: 29260260 Review.
-
Next-generation sequencing and protein mass spectrometry for the comprehensive analysis of human cellular and serum antibody repertoires.Curr Opin Chem Biol. 2015 Feb;24:112-20. doi: 10.1016/j.cbpa.2014.11.007. Epub 2014 Nov 25. Curr Opin Chem Biol. 2015. PMID: 25461729 Review.
-
Benchmarking and integrating human B-cell receptor genomic and antibody proteomic profiling.NPJ Syst Biol Appl. 2024 Jul 12;10(1):73. doi: 10.1038/s41540-024-00402-z. NPJ Syst Biol Appl. 2024. PMID: 38997321 Free PMC article.
Cited by
-
Systematic evaluation of B-cell clonal family inference approaches.BMC Immunol. 2024 Feb 8;25(1):13. doi: 10.1186/s12865-024-00600-8. BMC Immunol. 2024. PMID: 38331731 Free PMC article.
-
Current strategies for detecting functional convergence across B-cell receptor repertoires.MAbs. 2021 Jan-Dec;13(1):1996732. doi: 10.1080/19420862.2021.1996732. MAbs. 2021. PMID: 34781829 Free PMC article.
-
Computational Approaches and Challenges to Developing Universal Influenza Vaccines.Vaccines (Basel). 2019 May 28;7(2):45. doi: 10.3390/vaccines7020045. Vaccines (Basel). 2019. PMID: 31141933 Free PMC article. Review.
-
Novel metric for hyperbolic phylogenetic tree embeddings.Biol Methods Protoc. 2021 Mar 27;6(1):bpab006. doi: 10.1093/biomethods/bpab006. eCollection 2021. Biol Methods Protoc. 2021. PMID: 33928190 Free PMC article.
-
Mice carrying the full-length human immunoglobulin loci produce antigen-specific human antibodies with the lambda light chain.iScience. 2024 Oct 28;27(12):111258. doi: 10.1016/j.isci.2024.111258. eCollection 2024 Dec 20. iScience. 2024. PMID: 39758990 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

