Target RNAs Strike Back on MicroRNAs
- PMID: 30333855
- PMCID: PMC6175985
- DOI: 10.3389/fgene.2018.00435
Target RNAs Strike Back on MicroRNAs
Abstract
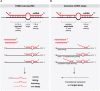
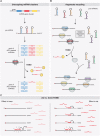
MicroRNAs are extensively studied regulatory non-coding small RNAs that silence animal genes throughout most biological processes, typically doing so by binding to partially complementary sequences within target RNAs. A plethora of studies has described detailed mechanisms for microRNA biogenesis and function, as well as their temporal and spatial regulation during development. By inducing translational repression and/or degradation of their target RNAs, microRNAs can contribute to achieve highly specific cell- or tissue-specific gene expression, while their aberrant expression can lead to disease. Yet an unresolved aspect of microRNA biology is how such small RNA molecules are themselves cleared from the cell, especially under circumstances where fast microRNA turnover or specific degradation of individual microRNAs is required. In recent years, it was unexpectedly found that binding of specific target RNAs to microRNAs with extensive complementarity can reverse the outcome, triggering degradation of the bound microRNAs. This emerging pathway, named TDMD for Target RNA-Directed MicroRNA Degradation, leads to microRNA 3'-end tailing by the addition of A/U non-templated nucleotides, trimming or shortening from the 3' end, and highly specific microRNA loss, providing a new layer of microRNA regulation. Originally described in flies and known to be triggered by viral RNAs, novel endogenous instances of TDMD have been uncovered and are now starting to be understood. Here, we review our current knowledge of this pathway and its potential role in the control and diversification of microRNA expression patterns.
Keywords: Argonaute; TDMD; degradation; exoribonuclease; microRNA; tailing and trimming; terminal nucleotidyl transferase; uridylation.
Figures


Similar articles
-
How Complementary Targets Expose the microRNA 3' End for Tailing and Trimming during Target-Directed microRNA Degradation.Cold Spring Harb Symp Quant Biol. 2019;84:179-183. doi: 10.1101/sqb.2019.84.039321. Epub 2020 Feb 4. Cold Spring Harb Symp Quant Biol. 2019. PMID: 32019864 Free PMC article.
-
Structural Basis for Target-Directed MicroRNA Degradation.Mol Cell. 2019 Sep 19;75(6):1243-1255.e7. doi: 10.1016/j.molcel.2019.06.019. Epub 2019 Jul 25. Mol Cell. 2019. PMID: 31353209 Free PMC article.
-
Target-directed microRNA degradation: Mechanisms, significance, and functional implications.Wiley Interdiscip Rev RNA. 2024 Mar-Apr;15(2):e1832. doi: 10.1002/wrna.1832. Wiley Interdiscip Rev RNA. 2024. PMID: 38448799 Free PMC article. Review.
-
MicroRNA turnover: a tale of tailing, trimming, and targets.Trends Biochem Sci. 2023 Jan;48(1):26-39. doi: 10.1016/j.tibs.2022.06.005. Epub 2022 Jul 7. Trends Biochem Sci. 2023. PMID: 35811249 Free PMC article. Review.
-
A ubiquitin ligase mediates target-directed microRNA decay independently of tailing and trimming.Science. 2020 Dec 18;370(6523):eabc9546. doi: 10.1126/science.abc9546. Epub 2020 Nov 12. Science. 2020. PMID: 33184234 Free PMC article.
Cited by
-
Enhancing arginase 2 expression using target site blockers as a strategy to modulate macrophage phenotype.Mol Ther Nucleic Acids. 2022 Aug 4;29:643-655. doi: 10.1016/j.omtn.2022.08.004. eCollection 2022 Sep 13. Mol Ther Nucleic Acids. 2022. PMID: 36090747 Free PMC article.
-
miRNA-based biomarkers, therapies, and resistance in Cancer.Int J Biol Sci. 2020 Jul 19;16(14):2628-2647. doi: 10.7150/ijbs.47203. eCollection 2020. Int J Biol Sci. 2020. PMID: 32792861 Free PMC article. Review.
-
Roles of circRNAs on tumor autophagy.Mol Ther Nucleic Acids. 2021 Jan 9;23:918-929. doi: 10.1016/j.omtn.2021.01.002. eCollection 2021 Mar 5. Mol Ther Nucleic Acids. 2021. PMID: 33614240 Free PMC article.
-
MiR-301b-3p promotes breast cancer development through inhibiting the expression of transforming growth factor-beta receptor 2.PeerJ. 2024 Nov 5;12:e18324. doi: 10.7717/peerj.18324. eCollection 2024. PeerJ. 2024. PMID: 39525474 Free PMC article.
-
Decoding microRNA arm switching: a key to evolutionary innovation and gene regulation.Cell Mol Life Sci. 2025 May 10;82(1):197. doi: 10.1007/s00018-025-05663-3. Cell Mol Life Sci. 2025. PMID: 40347284 Free PMC article. Review.
References
-
- Abe M., Yoshikawa T., Nosaka M., Sakakibara H., Sato Y., Nagato Y., et al. (2010). WAVY LEAF1, an ortholog of Arabidopsis HEN1, regulates shoot development by maintaining MicroRNA and trans-acting small interfering RNA accumulation in rice. Plant Physiol. 154 1335–1346. 10.1104/pp.110.160234 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources

