On the Importance of Host MicroRNAs During Viral Infection
- PMID: 30333857
- PMCID: PMC6176045
- DOI: 10.3389/fgene.2018.00439
On the Importance of Host MicroRNAs During Viral Infection
Abstract
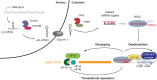
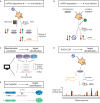
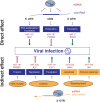
Every living organism has to constantly face threats from the environment and deal with a large number of pathogens against which it has to defend itself to survive. Among those, viruses represent a large class of obligatory intracellular parasites, which rely on their host machinery to multiply and propagate. As a result, viruses and their hosts have engaged in an ever-evolving arms race to be able to maintain their existence. The role played by micro (mi)RNAs in this ongoing battle has been extensively studied in the past 15 years and will be the subject of this review article. We will mainly focus on cellular miRNAs and their implication during viral infection in mammals. Thus, we will describe current techniques that can be used to identify miRNAs involved in the modulation of viral infection and to characterize their targets and mode of action. We will also present different reported examples of miRNA-mediated regulation of viruses, which can have a positive outcome either for the host or for the virus. In addition, the mode of action is also of a dual nature, depending on the target of the miRNA. Indeed, the regulatory small RNA can either directly guide an Argonaute protein on a viral transcript, or target a cellular mRNA involved in the host antiviral response. We will then see whether and how viruses respond to miRNA-mediated targeting. Finally, we will discuss how our knowledge of viral targeting by miRNA can be exploited for developing new antiviral therapeutic approaches.
Keywords: defense mechanism; host–pathogen interaction; microRNA; post-transcriptional regulation; virus.
Figures
Similar articles
-
Inducible MicroRNA-3570 Feedback Inhibits the RIG-I-Dependent Innate Immune Response to Rhabdovirus in Teleost Fish by Targeting MAVS/IPS-1.J Virol. 2018 Jan 2;92(2):e01594-17. doi: 10.1128/JVI.01594-17. Print 2018 Jan 15. J Virol. 2018. PMID: 29093090 Free PMC article.
-
Computational analysis of microRNA-mediated interactions in SARS-CoV-2 infection.PeerJ. 2020 Jun 5;8:e9369. doi: 10.7717/peerj.9369. eCollection 2020. PeerJ. 2020. PMID: 32547891 Free PMC article.
-
A Viral Suppressor Modulates the Plant Immune Response Early in Infection by Regulating MicroRNA Activity.mBio. 2018 Apr 24;9(2):e00419-18. doi: 10.1128/mBio.00419-18. mBio. 2018. PMID: 29691336 Free PMC article.
-
[Importance of cellular microRNAs in the regulation of viral infections].Med Sci (Paris). 2019 Aug-Sep;35(8-9):667-673. doi: 10.1051/medsci/2019130. Epub 2019 Sep 18. Med Sci (Paris). 2019. PMID: 31532379 Review. French.
-
MicroRNAs in virus-induced tumorigenesis and IFN system.Cytokine Growth Factor Rev. 2015 Apr;26(2):183-94. doi: 10.1016/j.cytogfr.2014.11.002. Epub 2014 Nov 13. Cytokine Growth Factor Rev. 2015. PMID: 25466647 Review.
Cited by
-
SARS-CoV-2 causes a significant stress response mediated by small RNAs in the blood of COVID-19 patients.Mol Ther Nucleic Acids. 2022 Mar 8;27:751-762. doi: 10.1016/j.omtn.2021.12.034. Epub 2021 Dec 31. Mol Ther Nucleic Acids. 2022. PMID: 35003892 Free PMC article.
-
Vitamin D modulates inflammatory response of DENV-2-infected macrophages by inhibiting the expression of inflammatory-liked miRNAs.Pathog Glob Health. 2023 Mar;117(2):167-180. doi: 10.1080/20477724.2022.2101840. Epub 2022 Jul 19. Pathog Glob Health. 2023. PMID: 35850625 Free PMC article.
-
The hidden RNA code: implications of the RNA epitranscriptome in the context of viral infections.Front Genet. 2023 Aug 1;14:1245683. doi: 10.3389/fgene.2023.1245683. eCollection 2023. Front Genet. 2023. PMID: 37614818 Free PMC article. Review.
-
Small non-coding RNAs encoded by RNA viruses: old controversies and new lessons from the COVID-19 pandemic.Front Genet. 2023 Jun 21;14:1216890. doi: 10.3389/fgene.2023.1216890. eCollection 2023. Front Genet. 2023. PMID: 37415603 Free PMC article. Review.
-
Integrative role of small non-coding RNAs in viral immune response: a systematic review.Mol Biol Rep. 2024 Jan 16;51(1):107. doi: 10.1007/s11033-023-09141-6. Mol Biol Rep. 2024. PMID: 38227137
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

