Crosstalk Between Mammalian Autophagy and the Ubiquitin-Proteasome System
- PMID: 30333975
- PMCID: PMC6175981
- DOI: 10.3389/fcell.2018.00128
Crosstalk Between Mammalian Autophagy and the Ubiquitin-Proteasome System
Abstract
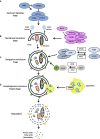
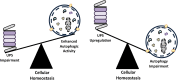
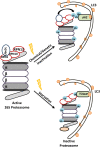
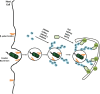
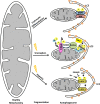
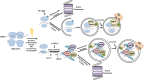
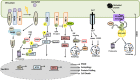
Autophagy and the ubiquitin-proteasome system (UPS) are the two major intracellular quality control and recycling mechanisms that are responsible for cellular homeostasis in eukaryotes. Ubiquitylation is utilized as a degradation signal by both systems, yet, different mechanisms are in play. The UPS is responsible for the degradation of short-lived proteins and soluble misfolded proteins whereas autophagy eliminates long-lived proteins, insoluble protein aggregates and even whole organelles (e.g., mitochondria, peroxisomes) and intracellular parasites (e.g., bacteria). Both the UPS and selective autophagy recognize their targets through their ubiquitin tags. In addition to an indirect connection between the two systems through ubiquitylated proteins, recent data indicate the presence of connections and reciprocal regulation mechanisms between these degradation pathways. In this review, we summarize these direct and indirect interactions and crosstalks between autophagy and the UPS, and their implications for cellular stress responses and homeostasis.
Keywords: UPS; autophagy; mitophagy; organelle homeostasis; proteasome; protein quality control; proteostasis; ubiquitylation.
Figures



References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

