Deep Geodesic Learning for Segmentation and Anatomical Landmarking
- PMID: 30334750
- PMCID: PMC6475529
- DOI: 10.1109/TMI.2018.2875814
Deep Geodesic Learning for Segmentation and Anatomical Landmarking
Abstract
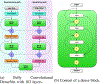
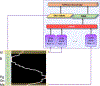
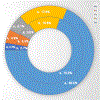
In this paper, we propose a novel deep learning framework for anatomy segmentation and automatic landmarking. Specifically, we focus on the challenging problem of mandible segmentation from cone-beam computed tomography (CBCT) scans and identification of 9 anatomical landmarks of the mandible on the geodesic space. The overall approach employs three inter-related steps. In the first step, we propose a deep neural network architecture with carefully designed regularization, and network hyper-parameters to perform image segmentation without the need for data augmentation and complex post-processing refinement. In the second step, we formulate the landmark localization problem directly on the geodesic space for sparsely-spaced anatomical landmarks. In the third step, we utilize a long short-term memory network to identify the closely-spaced landmarks, which is rather difficult to obtain using other standard networks. The proposed fully automated method showed superior efficacy compared to the state-of-the-art mandible segmentation and landmarking approaches in craniofacial anomalies and diseased states. We used a very challenging CBCT data set of 50 patients with a high-degree of craniomaxillofacial variability that is realistic in clinical practice. The qualitative visual inspection was conducted for distinct CBCT scans from 250 patients with high anatomical variability. We have also shown the state-of-the-art performance in an independent data set from the MICCAI Head-Neck Challenge (2015).
Figures
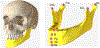
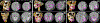
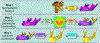


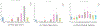
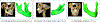


References
-
- Armond ACV, Martins C, Glria J, Galvo E, dos Santos C, and Falci S, “Influence of third molars in mandibular fractures. part 1: mandibular anglea meta-analysis,” International Journal of Oral and Maxillofacial Surgery, vol. 46, no. 6, pp. 716–729, 2017. - PubMed
-
- Anuwongnukroh N, Dechkunakorn S, Damrongsri S, Nilwarat C, Pudpong N, Radomsutthisarn W, and Kangern S, “Accuracy of automatic cephalometric software on landmark identification,” IOP Conference Series: Materials Science and Engineering, vol. 265, no. 1, p. 012028, 2017.
-
- Zhang J, Gao Y, Wang L, Tang Z, Xia JJ, and Shen D, “Automatic craniomaxillofacial landmark digitization via segmentation-guided partially-joint regression forest model and multiscale statistical features,” IEEE Transactions on Biomedical Engineering, vol. 63, no. 9, pp. 1820–1829, September 2016. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

