Androgen receptor splice variant-7 expression emerges with castration resistance in prostate cancer
- PMID: 30334814
- PMCID: PMC6307949
- DOI: 10.1172/JCI122819
Androgen receptor splice variant-7 expression emerges with castration resistance in prostate cancer
Abstract
Background: Liquid biopsies have demonstrated that the constitutively active androgen receptor splice variant-7 (AR-V7) associates with reduced response and overall survival from endocrine therapies in castration-resistant prostate cancer (CRPC). However, these studies provide little information pertaining to AR-V7 expression in prostate cancer (PC) tissue.
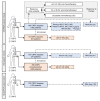
Methods: Following generation and validation of a potentially novel AR-V7 antibody for IHC, AR-V7 protein expression was determined for 358 primary prostate samples and 293 metastatic biopsies. Associations with disease progression, full-length androgen receptor (AR-FL) expression, response to therapy, and gene expression were determined.
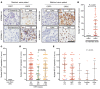
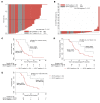
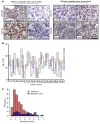
Results: We demonstrated that AR-V7 protein is rarely expressed (<1%) in primary PC but is frequently detected (75% of cases) following androgen deprivation therapy, with further significant (P = 0.020) increase in expression following abiraterone acetate or enzalutamide therapy. In CRPC, AR-V7 expression is predominantly (94% of cases) nuclear and correlates with AR-FL expression (P ≤ 0.001) and AR copy number (P = 0.026). However, dissociation of expression was observed, suggesting that mRNA splicing remains crucial for AR-V7 generation. AR-V7 expression was heterogeneous between different metastases from a patient, although AR-V7 expression was similar within a metastasis. Moreover, AR-V7 expression correlated with a unique 59-gene signature in CRPC, including HOXB13, a critical coregulator of AR-V7 function. Finally, AR-V7-negative disease associated with better prostate-specific antigen (PSA) responses (100% vs. 54%, P = 0.03) and overall survival (74.3 vs. 25.2 months, hazard ratio 0.23 [0.07-0.79], P = 0.02) from endocrine therapies (pre-chemotherapy).
Conclusion: This study provides impetus to develop therapies that abrogate AR-V7 signaling to improve our understanding of AR-V7 biology and to confirm the clinical significance of AR-V7.
Funding: Work at the University of Washington and in the Plymate and Nelson laboratories is supported by the Department of Defense Prostate Cancer Research Program (W81XWH-14-2-0183, W81XWH-12-PCRP-TIA, W81XWH-15-1-0430, and W81XWH-13-2-0070), the Pacific Northwest Prostate Cancer SPORE (P50CA97186), the Institute for Prostate Cancer Research, the Veterans Affairs Research Program, the NIH/National Cancer Institute (P01CA163227), and the Prostate Cancer Foundation. Work in the de Bono laboratory was supported by funding from the Movember Foundation/Prostate Cancer UK (CEO13-2-002), the US Department of Defense (W81XWH-13-2-0093), the Prostate Cancer Foundation (20131017 and 20131017-1), Stand Up To Cancer (SU2C-AACR-DT0712), Cancer Research UK (CRM108X-A25144), and the UK Department of Health through an Experimental Cancer Medicine Centre grant (ECMC-CRM064X).
Keywords: Oncology; Prostate cancer.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

