The mouse Gene Expression Database (GXD): 2019 update
- PMID: 30335138
- PMCID: PMC6324054
- DOI: 10.1093/nar/gky922
The mouse Gene Expression Database (GXD): 2019 update
Abstract
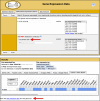
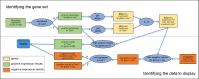
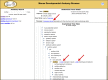
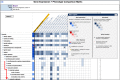
The mouse Gene Expression Database (GXD) is an extensive, well-curated community resource freely available at www.informatics.jax.org/expression.shtml. Covering all developmental stages, GXD includes data from RNA in situ hybridization, immunohistochemistry, RT-PCR, northern blot and western blot experiments in wild-type and mutant mice. GXD's gene expression information is integrated with the other data in Mouse Genome Informatics and interconnected with other databases, placing these data in the larger biological and biomedical context. Since the last report, the ability of GXD to provide insights into the molecular mechanisms of development and disease has been greatly enhanced by the addition of new data and by the implementation of new web features. These include: improvements to the Differential Gene Expression Data Search, facilitating searches for genes that have been shown to be exclusively expressed in a specified structure and/or developmental stage; an enhanced anatomy browser that now provides access to expression data and phenotype data for a given anatomical structure; direct access to the wild-type gene expression data for the tissues affected in a specific mutant; and a comparison matrix that juxtaposes tissues where a gene is normally expressed against tissues, where mutations in that gene cause abnormalities.
Figures
References
-
- Ringwald M., Baldock R., Bard J., Kaufman M., Eppig J.T., Richardson J.E., Nadeau J.H., Davidson D.. A database for mouse development. Science. 1994; 265:2033–2034. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

