Comprehensive analysis of long noncoding RNA expression in dorsal root ganglion reveals cell-type specificity and dysregulation after nerve injury
- PMID: 30335683
- PMCID: PMC6343954
- DOI: 10.1097/j.pain.0000000000001416
Comprehensive analysis of long noncoding RNA expression in dorsal root ganglion reveals cell-type specificity and dysregulation after nerve injury
Abstract
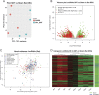
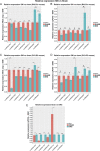
Dorsal root ganglion (DRG) neurons provide connectivity between peripheral tissues and the spinal cord. Transcriptional plasticity within DRG sensory neurons after peripheral nerve injury contributes to nerve repair but also leads to maladaptive plasticity, including the development of neuropathic pain. This study presents tissue and neuron-specific expression profiling of both known and novel long noncoding RNAs (LncRNAs) in the rodent DRG after nerve injury. We have identified a large number of novel LncRNAs expressed within the rodent DRG, a minority of which were syntenically conserved between the mouse, rat, and human, and including, both intergenic and antisense LncRNAs. We have also identified neuron type-specific LncRNAs in the mouse DRG and LncRNAs that are expressed in human IPS cell-derived sensory neurons. We show significant plasticity in LncRNA expression after nerve injury, which in mice is strain and gender dependent. This resource is publicly available and will aid future studies of DRG neuron identity and the transcriptional landscape in both the naive and injured DRG. We present our work regarding novel antisense and intergenic LncRNAs as an online searchable database, accessible from PainNetworks (http://www.painnetworks.org/). We have also integrated all annotated gene expression data in PainNetworks, so they can be examined in the context of their protein interactions.
Conflict of interest statement
Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article.
Figures




References
-
- Alexa A, Rahnenfuhrer J. topGO: topGO: enrichment analysis for gene ontology. 2010. www.bioconductor.org. In R package version 3.4.4.
-
- van Brakel JP. Algorithm—peak signal detection in realtime timeseries data—stack overflow. 2014. Available at: https://stackoverflow.com/questions/22583391/peak-signal-detection-in-re.... Accessed May 1, 2018.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

