Modeling large fluctuations of thousands of clones during hematopoiesis: The role of stem cell self-renewal and bursty progenitor dynamics in rhesus macaque
- PMID: 30335762
- PMCID: PMC6218102
- DOI: 10.1371/journal.pcbi.1006489
Modeling large fluctuations of thousands of clones during hematopoiesis: The role of stem cell self-renewal and bursty progenitor dynamics in rhesus macaque
Abstract
In a recent clone-tracking experiment, millions of uniquely tagged hematopoietic stem cells (HSCs) and progenitor cells were autologously transplanted into rhesus macaques and peripheral blood containing thousands of tags were sampled and sequenced over 14 years to quantify the abundance of hundreds to thousands of tags or "clones." Two major puzzles of the data have been observed: consistent differences and massive temporal fluctuations of clone populations. The large sample-to-sample variability can lead clones to occasionally go "extinct" but "resurrect" themselves in subsequent samples. Although heterogeneity in HSC differentiation rates, potentially due to tagging, and random sampling of the animals' blood and cellular demographic stochasticity might be invoked to explain these features, we show that random sampling cannot explain the magnitude of the temporal fluctuations. Moreover, we show through simpler neutral mechanistic and statistical models of hematopoiesis of tagged cells that a broad distribution in clone sizes can arise from stochastic HSC self-renewal instead of tag-induced heterogeneity. The very large clone population fluctuations that often lead to extinctions and resurrections can be naturally explained by a generation-limited proliferation constraint on the progenitor cells. This constraint leads to bursty cell population dynamics underlying the large temporal fluctuations. We analyzed experimental clone abundance data using a new statistic that counts clonal disappearances and provided least-squares estimates of two key model parameters in our model, the total HSC differentiation rate and the maximum number of progenitor-cell divisions.
Conflict of interest statement
Dr. Irvin S. Y. Chen has a financial interest in CSL Behring and Calimmune Inc. No funding was provided by these companies to support this work.
Figures


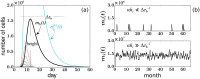
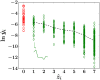
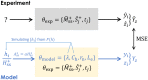
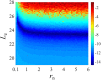




Similar articles
-
Mechanisms of blood homeostasis: lineage tracking and a neutral model of cell populations in rhesus macaques.BMC Biol. 2015 Oct 20;13:85. doi: 10.1186/s12915-015-0191-8. BMC Biol. 2015. PMID: 26486451 Free PMC article.
-
Quantitative stability of hematopoietic stem and progenitor cell clonal output in rhesus macaques receiving transplants.Blood. 2017 Mar 16;129(11):1448-1457. doi: 10.1182/blood-2016-07-728691. Epub 2017 Jan 13. Blood. 2017. PMID: 28087539 Free PMC article.
-
Clonal-level lineage commitment pathways of hematopoietic stem cells in vivo.Proc Natl Acad Sci U S A. 2019 Jan 22;116(4):1447-1456. doi: 10.1073/pnas.1801480116. Epub 2019 Jan 8. Proc Natl Acad Sci U S A. 2019. PMID: 30622181 Free PMC article.
-
Aging and Clonal Behavior of Hematopoietic Stem Cells.Int J Mol Sci. 2022 Feb 9;23(4):1948. doi: 10.3390/ijms23041948. Int J Mol Sci. 2022. PMID: 35216063 Free PMC article. Review.
-
Renewal and release of hemopoietic stem cells: does clonal succession exist?Blood Cells. 1986;12(1):103-27. Blood Cells. 1986. PMID: 3539233 Review.
Cited by
-
Gene therapy using haematopoietic stem and progenitor cells.Nat Rev Genet. 2021 Apr;22(4):216-234. doi: 10.1038/s41576-020-00298-5. Epub 2020 Dec 10. Nat Rev Genet. 2021. PMID: 33303992 Review.
-
Evolution of cancer stem cell lineage involving feedback regulation.PLoS One. 2021 May 20;16(5):e0251481. doi: 10.1371/journal.pone.0251481. eCollection 2021. PLoS One. 2021. PMID: 34014979 Free PMC article.
-
In vivo dynamics of human hematopoietic stem cells: novel concepts and future directions.Blood Adv. 2019 Jun 25;3(12):1916-1924. doi: 10.1182/bloodadvances.2019000039. Blood Adv. 2019. PMID: 31239246 Free PMC article. Review.
-
Stem cell mutations, associated cancer risk, and consequences for regenerative medicine.Cell Stem Cell. 2023 Nov 2;30(11):1421-1433. doi: 10.1016/j.stem.2023.09.008. Epub 2023 Oct 12. Cell Stem Cell. 2023. PMID: 37832550 Free PMC article. Review.
-
Dynamically adjusted cell fate decisions and resilience to mutant invasion during steady-state hematopoiesis revealed by an experimentally parameterized mathematical model.Proc Natl Acad Sci U S A. 2024 Sep 17;121(38):e2321525121. doi: 10.1073/pnas.2321525121. Epub 2024 Sep 9. Proc Natl Acad Sci U S A. 2024. PMID: 39250660 Free PMC article.

