Enzymatic Transition States and Drug Design
- PMID: 30335982
- PMCID: PMC6615489
- DOI: 10.1021/acs.chemrev.8b00369
Enzymatic Transition States and Drug Design
Abstract
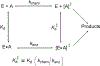
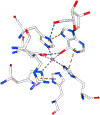
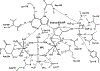
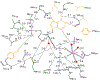
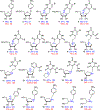
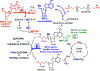
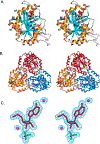
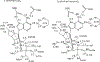
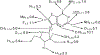
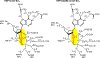
Transition state theory teaches that chemically stable mimics of enzymatic transition states will bind tightly to their cognate enzymes. Kinetic isotope effects combined with computational quantum chemistry provides enzymatic transition state information with sufficient fidelity to design transition state analogues. Examples are selected from various stages of drug development to demonstrate the application of transition state theory, inhibitor design, physicochemical characterization of transition state analogues, and their progress in drug development.
Conflict of interest statement
The author declares the following competing financial interest(s): The author serves as a consultant or board member to several biotech companies involved in development of the immucillin family of transition state analogues, some of them described here. The intellectual property developed around these inhibitors is the joint property of the Albert Einstein College of Medicine and Victoria University of Wellington, New Zealand. The author and collaborators receive royalties through dividend plans of these institutions.
Figures


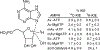

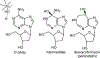
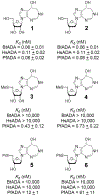
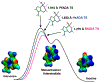
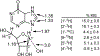
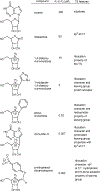

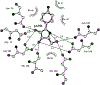

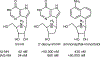
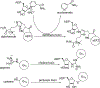

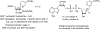




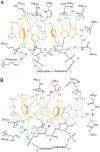

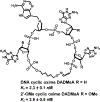
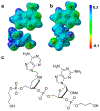
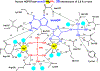





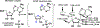

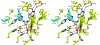

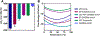
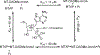
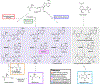
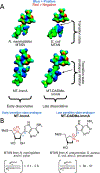










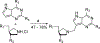
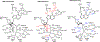


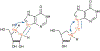
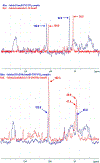
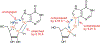
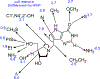

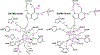

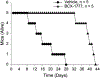
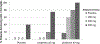


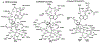


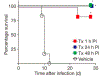

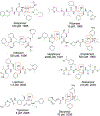
References
-
- Pauling L Molecular Architecture and Biological Reactions. Chem. Eng. News 1946, 24, 1375–1377.
-
- Pauling L Nature of Forces Between Large Molecules of Biological Interest. Nature 1948, 161, 707–709. - PubMed
-
- Wolfenden R Transition State Analogues for Enzyme Catalysis. Nature 1969, 223, 704–705. - PubMed
-
- Wolfenden R Transition State Analog Inhibitors and Enzyme Catalysis. Annu. Rev. Biophys. Bioeng 1976, 5, 271–306. - PubMed
-
- Wolfenden R Thermodynamic and Extrathermodynamic Requirements of Enzyme Catalysis. Biophys. Chem 2003, 105, 559–572. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

