The Complete Plastome Sequences of Eleven Capsicum Genotypes: Insights into DNA Variation and Molecular Evolution
- PMID: 30336638
- PMCID: PMC6210379
- DOI: 10.3390/genes9100503
The Complete Plastome Sequences of Eleven Capsicum Genotypes: Insights into DNA Variation and Molecular Evolution
Abstract
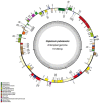
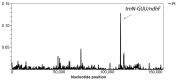
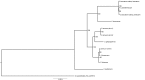
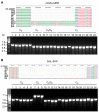
Members of the genus Capsicum are of great economic importance, including both wild forms and cultivars of peppers and chilies. The high number of potentially informative characteristics that can be identified through next-generation sequencing technologies gave a huge boost to evolutionary and comparative genomic research in higher plants. Here, we determined the complete nucleotide sequences of the plastomes of eight Capsicum species (eleven genotypes), representing the three main taxonomic groups in the genus and estimated molecular diversity. Comparative analyses highlighted a wide spectrum of variation, ranging from point mutations to small/medium size insertions/deletions (InDels), with accD, ndhB, rpl20, ycf1, and ycf2 being the most variable genes. The global pattern of sequence variation is consistent with the phylogenetic signal. Maximum-likelihood tree estimation revealed that Capsicum chacoense is sister to the baccatum complex. Divergence and positive selection analyses unveiled that protein-coding genes were generally well conserved, but we identified 25 positive signatures distributed in six genes involved in different essential plastid functions, suggesting positive selection during evolution of Capsicum plastomes. Finally, the identified sequence variation allowed us to develop simple PCR-based markers useful in future work to discriminate species belonging to different Capsicum complexes.
Keywords: chloroplast genome; microsatellites; molecular markers; next-generation sequencing; pepper; perfect tandem repeats; sequence variability; simple sequence repeats; single-nucleotide polymorphism.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Jansen R.K., Cai Z., Raubeson L.A., Daniell H., de Pamphilis C.W., Leebens-Mack J., Müller K.F., Guisinger-Bellian M., Haberle R.C., Hansen A.K., et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in Angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. USA. 2007;104:19369–19374. doi: 10.1073/pnas.0709121104. - DOI - PMC - PubMed
-
- Olmstead R.G., Bohs L., Migid H.A., Santiago-Valentin E., Garcia V.F., Collier S.M. A molecular phylogeny of the Solanaceae. Taxon. 2008;57:1159–1181.
-
- Chase M.W., Christenhusz M.J.M., Fay M.F., Byng J.W., Judd W.S., Soltis D.E., Mabberley D.J., Sennikov A.N., Soltis P.S., Stevens P.F., et al. An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016;181:1–20.
-
- Cheng J., Zhao Z., Li B., Qin C., Wu Z., Trejo-Saavedra D.L., Luo X., Cui J., Rivera-Bustamante R.F., Li S., et al. A comprehensive characterization of simple sequence repeats in pepper genomes provides valuable resources for marker development in Capsicum. Sci. Rep. 2016;6:18919. doi: 10.1038/srep18919. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

