Consistent responses of soil microbial taxonomic and functional attributes to mercury pollution across China
- PMID: 30336790
- PMCID: PMC6194565
- DOI: 10.1186/s40168-018-0572-7
Consistent responses of soil microbial taxonomic and functional attributes to mercury pollution across China
Abstract
Background: The ecological consequences of mercury (Hg) pollution-one of the major pollutants worldwide-on microbial taxonomic and functional attributes remain poorly understood and largely unexplored. Using soils from two typical Hg-impacted regions across China, here, we evaluated the role of Hg pollution in regulating bacterial abundance, diversity, and co-occurrence network. We also investigated the associations between Hg contents and the relative abundance of microbial functional genes by analyzing the soil metagenomes from a subset of those sites.
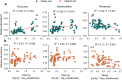
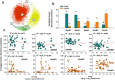
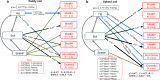
Results: We found that soil Hg largely influenced the taxonomic and functional attributes of microbial communities in the two studied regions. In general, Hg pollution was negatively related to bacterial abundance, but positively related to the diversity of bacteria in two separate regions. We also found some consistent associations between soil Hg contents and the community composition of bacteria. For example, soil total Hg content was positively related to the relative abundance of Firmicutes and Bacteroidetes in both paddy and upland soils. In contrast, the methylmercury (MeHg) concentration was negatively correlated to the relative abundance of Nitrospirae in the two types of soils. Increases in soil Hg pollution correlated with drastic changes in the relative abundance of ecological clusters within the co-occurrence network of bacterial communities for the two regions. Using metagenomic data, we were also able to detect the effect of Hg pollution on multiple functional genes relevant to key soil processes such as element cycles and Hg transformations (e.g., methylation and reduction).
Conclusions: Together, our study provides solid evidence that Hg pollution has predictable and significant effects on multiple taxonomic and functional attributes including bacterial abundance, diversity, and the relative abundance of ecological clusters and functional genes. Our results suggest an increase in soil Hg pollution linked to human activities will lead to predictable shifts in the taxonomic and functional attributes in the Hg-impacted areas, with potential implications for sustainable management of agricultural ecosystems and elsewhere.
Keywords: Co-occurrence network; Functional gene; Mercury pollution; Metagenomics; Soil microbial community.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

