Notch signalling patterns retinal composition by regulating atoh7 during post-embryonic growth
- PMID: 30337377
- PMCID: PMC6240314
- DOI: 10.1242/dev.169698
Notch signalling patterns retinal composition by regulating atoh7 during post-embryonic growth
Abstract
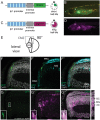
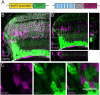
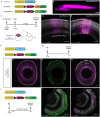
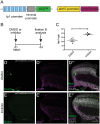
Patterning of a continuously growing naive field in the context of a life-long growing organ such as the teleost eye is of high functional relevance. Intrinsic and extrinsic signals have been proposed to regulate lineage specification in progenitors that exit the stem cell niche in the ciliary marginal zone (CMZ). The proper cell-type composition arising from those progenitors is a prerequisite for retinal function. Our findings in the teleost medaka (Oryzias latipes) uncover that the Notch-Atoh7 axis continuously patterns the CMZ. The complement of cell types originating from the two juxtaposed progenitors marked by Notch or Atoh7 activity contains all constituents of a retinal column. Modulation of Notch signalling specifically in Atoh7-expressing cells demonstrates the crucial role of this axis in generating the correct cell-type proportions. After transiently blocking Notch signalling, retinal patterning and differentiation is re-initiated de novo Taken together, our data show that Notch activity in the CMZ continuously structures the growing retina by juxtaposing Notch and Atoh7 progenitors that give rise to distinct complementary lineages, revealing coupling of de novo patterning and cell-type specification in the respective lineages.
Keywords: Atoh7; Cell specification; Medaka; Notch; Post-embryonic growth; Retinal progenitors.
© 2018. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures


Similar articles
-
Retinal Stem Cell 'Retirement Plans': Growth, Regulation and Species Adaptations in the Retinal Ciliary Marginal Zone.Int J Mol Sci. 2021 Jun 18;22(12):6528. doi: 10.3390/ijms22126528. Int J Mol Sci. 2021. PMID: 34207050 Free PMC article. Review.
-
Reprogramming amacrine and photoreceptor progenitors into retinal ganglion cells by replacing Neurod1 with Atoh7.Development. 2013 Feb 1;140(3):541-51. doi: 10.1242/dev.085886. Development. 2013. PMID: 23293286 Free PMC article.
-
Gene expression is dynamically regulated in retinal progenitor cells prior to and during overt cellular differentiation.Gene Expr Patterns. 2014 Jan;14(1):42-54. doi: 10.1016/j.gep.2013.10.003. Epub 2013 Oct 19. Gene Expr Patterns. 2014. PMID: 24148613
-
De novo neurogenesis by targeted expression of atoh7 to Müller glia cells.Development. 2016 Jun 1;143(11):1874-83. doi: 10.1242/dev.135905. Epub 2016 Apr 11. Development. 2016. PMID: 27068106 Free PMC article.
-
Intrinsic control of mammalian retinogenesis.Cell Mol Life Sci. 2013 Jul;70(14):2519-32. doi: 10.1007/s00018-012-1183-2. Epub 2012 Oct 12. Cell Mol Life Sci. 2013. PMID: 23064704 Free PMC article. Review.
Cited by
-
Igf signaling couples retina growth with body growth by modulating progenitor cell division.Development. 2021 Apr 1;148(7):dev199133. doi: 10.1242/dev.199133. Epub 2021 Apr 15. Development. 2021. PMID: 33722901 Free PMC article.
-
Retinal Stem Cell 'Retirement Plans': Growth, Regulation and Species Adaptations in the Retinal Ciliary Marginal Zone.Int J Mol Sci. 2021 Jun 18;22(12):6528. doi: 10.3390/ijms22126528. Int J Mol Sci. 2021. PMID: 34207050 Free PMC article. Review.
-
Differential Responses of Neural Retina Progenitor Populations to Chronic Hyperglycemia.Cells. 2021 Nov 22;10(11):3265. doi: 10.3390/cells10113265. Cells. 2021. PMID: 34831487 Free PMC article.
-
Cell Sources for Retinal Regeneration: Implication for Data Translation in Biomedicine of the Eye.Cells. 2022 Nov 24;11(23):3755. doi: 10.3390/cells11233755. Cells. 2022. PMID: 36497013 Free PMC article. Review.
-
αβ/γδ T cell lineage outcome is regulated by intrathymic cell localization and environmental signals.Sci Adv. 2021 Jul 14;7(29):eabg3613. doi: 10.1126/sciadv.abg3613. Print 2021 Jul. Sci Adv. 2021. PMID: 34261656 Free PMC article.
References
-
- Austin C. P., Feldman D. E., Ida J. A. and Cepko C. L. (1995). Vertebrate retinal ganglion cells are selected from competent progenitors by the action of Notch. Development 121, 3637-3650. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

