The dark side of centromeres: types, causes and consequences of structural abnormalities implicating centromeric DNA
- PMID: 30337534
- PMCID: PMC6194107
- DOI: 10.1038/s41467-018-06545-y
The dark side of centromeres: types, causes and consequences of structural abnormalities implicating centromeric DNA
Abstract
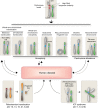
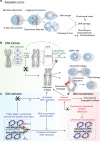
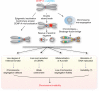
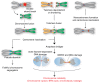
Centromeres are the chromosomal domains required to ensure faithful transmission of the genome during cell division. They have a central role in preventing aneuploidy, by orchestrating the assembly of several components required for chromosome separation. However, centromeres also adopt a complex structure that makes them susceptible to being sites of chromosome rearrangements. Therefore, preservation of centromere integrity is a difficult, but important task for the cell. In this review, we discuss how centromeres could potentially be a source of genome instability and how centromere aberrations and rearrangements are linked with human diseases such as cancer.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

