smarce1 mutants have a defective endocardium and an increased expression of cardiac transcription factors in zebrafish
- PMID: 30337622
- PMCID: PMC6194089
- DOI: 10.1038/s41598-018-33746-8
smarce1 mutants have a defective endocardium and an increased expression of cardiac transcription factors in zebrafish
Abstract
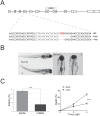
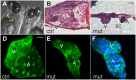
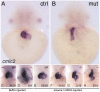
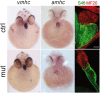
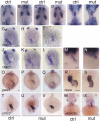
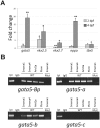
SWI/SNF or BAF chromatin-remodeling complexes are polymorphic assemblies of homologous subunit families that remodel nucleosomes and facilitate tissue-specific gene regulation during development. BAF57/SMARCE1 is a BAF complex subunit encoded in animals by a single gene and is a component of all mammalian BAF complexes. In vivo, the loss of SMARCE1 would lead to the formation of deficient combinations of the complex which might present limited remodeling activities. To address the specific contribution of SMARCE1 to the function of the BAF complex, we generated CRISPR/Cas9 mutations of smarce1 in zebrafish. Smarce1 mutants showed visible defects at 72 hpf, including smaller eyes, abnormal body curvature and heart abnormalities. Gene expression analysis revealed that the mutant embryos displayed defects in endocardial development since early stages, which led to the formation of a misshapen heart tube. The severe morphological and functional cardiac problems observed at 4 dpf were correlated with the substantially increased expression of different cardiac transcription factors. Additionally, we showed that Smarce1 binds to cis-regulatory regions of the gata5 gene and is necessary for the recruitment of the BAF complex to these regions.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

