Modified SureSelectQXT Target Enrichment Protocol for Illumina Multiplexed Sequencing of FFPE Samples
- PMID: 30337841
- PMCID: PMC6182866
- DOI: 10.1186/s12575-018-0084-7
Modified SureSelectQXT Target Enrichment Protocol for Illumina Multiplexed Sequencing of FFPE Samples
Abstract
Background: Personalised medicine is nowadays a major objective in oncology. Molecular characterization of tumours through NGS offers the possibility to find possible therapeutic targets in a time- and cost-effective way. However, the low quality and complexity of FFPE DNA samples bring a series of disadvantages for massive parallel sequencing techniques compared to high-quality DNA samples (from blood cells, cell cultures, etc.).
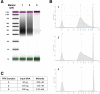
Results: We performed several experiments to understand the behaviour of FFPE DNA samples during the construction of SureSelectQXT libraries. First, we designed a quality checkpoint for FFPE DNA samples based on the quantification of their amplification capability (qcPCR). We observed that FFPE DNA samples can be classified according to DIN value and qcPCR concentration into unusable, or low-quality (LQ) and good-quality (GQ) DNA. For GQ samples, we increased the amount of input DNA to 150 ng and the digestion time to 30 min, whereas for LQ samples, we used 50 ng of DNA as input but we decreased the digestion time to 1 min. In all cases, we increased the cycles of the pre-hyb PCR to 10 but decreased the cycles of the post-hyb PCR to 8. In addition, we confirmed that using half of the volume of reagents can be beneficial. Finally, in order to obtain better results, we designed a decision flow-chart to achieve a seeding concentration of 12-14 pM for MiSeq Reagent Kit v2.
Conclusions: Our experiments allowed us to unveil the behaviour of low-quality FFPE DNA samples during the construction of SureSelectQXT libraries. Sequencing results showed that, using our modified SureSelectQXT protocol, the final percentage of usable reads for low-quality samples was increased more than three times allowing to reach median depth/million reads values of 76.35. This value is equivalent to ~ 0.9 and ~ 0.7 of the values obtained for good-quality FFPE and high-quality DNA respectively.
Keywords: FFPE samples; NGS; Optimization; Protocol; SureSelectQXT.
Conflict of interest statement
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. For this type of study formal consent is not required.Not applicableThe authors declare that they have no competing interests.Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures





References
LinkOut - more resources
Full Text Sources

