Calculation of Single Cell Assimilation Rates From SIP-NanoSIMS-Derived Isotope Ratios: A Comprehensive Approach
- PMID: 30337916
- PMCID: PMC6178922
- DOI: 10.3389/fmicb.2018.02342
Calculation of Single Cell Assimilation Rates From SIP-NanoSIMS-Derived Isotope Ratios: A Comprehensive Approach
Abstract
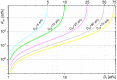
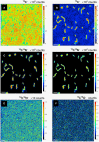
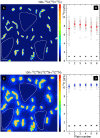
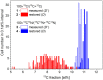
The nanoSIMS-based chemical microscopy has been introduced in biology over a decade ago. The spatial distribution of elements and isotopes analyzed by nanoSIMS can be used to reconstruct images of biological samples with a resolution down to tens of nanometers, and can be also interpreted quantitatively. Currently, a unified approach for calculation of single cell assimilation rates from nanoSIMS-derived changes in isotope ratios is missing. Here we present a comprehensive concept of assimilation rate calculation with a rigorous mathematical model based on quantitative evaluation of nanoSIMS-derived isotope ratios. We provide a detailed description of data acquisition and treatment, including the selection and accumulation of nanoSIMS scans, defining regions of interest and extraction of isotope ratios. Next, we present alternative methods to determine the cellular volume and the density of the element under scrutiny. Finally, to compensate for alterations of original isotopic ratios, our model considers corrections for sample preparation methods (e.g., air dry, chemical fixation, permeabilization, hybridization), and when known, for the stable isotope fractionation associated with utilization of defined growth substrates. As proof of concept we implemented this protocol to quantify the assimilation of 13C-labeled glucose by single cells of Pseudomonas putida. In addition, we provide a calculation template where all protocol-derived formulas are directly available to facilitate routine assimilation rate calculations by nanoSIMS users.
Keywords: assimilation rate; functional heterogeneity; isotope fractionation; isotope ratio; nanoSIMS; single cell; stable isotope probing.
Figures
References
-
- Fagerbakke K. M., Heldal M., Norland S. (1996). Content of carbon, nitrogen, oxygen, sulfur and phosphorus in native aquatic and cultured bacteria. Aquat. Microb. Ecol. 10, 15–27. 10.3354/ame010015 - DOI
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

