The Landscape of Repetitive Elements in the Refined Genome of Chilli Anthracnose Fungus Colletotrichum truncatum
- PMID: 30337918
- PMCID: PMC6180176
- DOI: 10.3389/fmicb.2018.02367
The Landscape of Repetitive Elements in the Refined Genome of Chilli Anthracnose Fungus Colletotrichum truncatum
Abstract
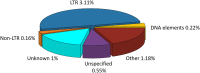
The ascomycete fungus Colletotrichum truncatum is a major phytopathogen with a broad host range which causes anthracnose disease of chilli. The genome sequencing of this fungus led to the discovery of functional categories of genes that may play important roles in fungal pathogenicity. However, the presence of gaps in C. truncatum draft assembly prevented the accurate prediction of repetitive elements, which are the key players to determine the genome architecture and drive evolution and host adaptation. We re-sequenced its genome using single-molecule real-time (SMRT) sequencing technology to obtain a refined assembly with lesser and smaller gaps and ambiguities. This enabled us to study its genome architecture by characterising the repetitive sequences like transposable elements (TEs) and simple sequence repeats (SSRs), which constituted 4.9 and 0.38% of the assembled genome, respectively. The comparative analysis among different Colletotrichum species revealed the extensive repeat rich regions, dominated by Gypsy superfamily of long terminal repeats (LTRs), and the differential composition of SSRs in their genomes. Our study revealed a recent burst of LTR amplification in C. truncatum, C. higginsianum, and C. scovillei. TEs in C. truncatum were significantly associated with secretome, effectors and genes in secondary metabolism clusters. Some of the TE families in C. truncatum showed cytosine to thymine transitions indicative of repeat-induced point mutation (RIP). C. orbiculare and C. graminicola showed strong signatures of RIP across their genomes and "two-speed" genomes with extensive AT-rich and gene-sparse regions. Comparative genomic analyses of Colletotrichum species provided an insight into the species-specific SSR profiles. The SSRs in the coding and non-coding regions of the genome revealed the composition of trinucleotide repeat motifs in exons with potential to alter the translated protein structure through amino acid repeats. This is the first genome-wide study of TEs and SSRs in C. truncatum and their comparative analysis with six other Colletotrichum species, which would serve as a useful resource for future research to get insights into the potential role of TEs in genome expansion and evolution of Colletotrichum fungi and for development of SSR-based molecular markers for population genomic studies.
Keywords: Colletotrichum truncatum; comparative genomics; repetitive DNA sequences; simple sequence repeats (SSRs); transposable elements (TEs); whole genome sequence.
Figures
Similar articles
-
Genome sequencing and comparative genomics reveal a repertoire of putative pathogenicity genes in chilli anthracnose fungus Colletotrichum truncatum.PLoS One. 2017 Aug 28;12(8):e0183567. doi: 10.1371/journal.pone.0183567. eCollection 2017. PLoS One. 2017. PMID: 28846714 Free PMC article.
-
Gapless genome assembly of Colletotrichum higginsianum reveals chromosome structure and association of transposable elements with secondary metabolite gene clusters.BMC Genomics. 2017 Aug 29;18(1):667. doi: 10.1186/s12864-017-4083-x. BMC Genomics. 2017. PMID: 28851275 Free PMC article.
-
The landscape of transposable elements in the finished genome of the fungal wheat pathogen Mycosphaerella graminicola.BMC Genomics. 2014 Dec 17;15(1):1132. doi: 10.1186/1471-2164-15-1132. BMC Genomics. 2014. PMID: 25519841 Free PMC article.
-
Colletotrichum higginsianum as a Model for Understanding Host⁻Pathogen Interactions: A Review.Int J Mol Sci. 2018 Jul 23;19(7):2142. doi: 10.3390/ijms19072142. Int J Mol Sci. 2018. PMID: 30041456 Free PMC article. Review.
-
The Hidden Truths of Fungal Virulence and Adaptation on Hosts: Unraveling the Conditional Dispensability of Minichromosomes in the Hemibiotrophic Colletotrichum Pathogens.Int J Mol Sci. 2023 Dec 22;25(1):198. doi: 10.3390/ijms25010198. Int J Mol Sci. 2023. PMID: 38203369 Free PMC article. Review.
Cited by
-
A practical comparison of the next-generation sequencing platform and assemblers using yeast genome.Life Sci Alliance. 2023 Feb 6;6(4):e202201744. doi: 10.26508/lsa.202201744. Print 2023 Apr. Life Sci Alliance. 2023. PMID: 36746534 Free PMC article.
-
The landscape and predicted roles of structural variants in Fusarium graminearum genomes.G3 (Bethesda). 2024 Jun 5;14(6):jkae065. doi: 10.1093/g3journal/jkae065. G3 (Bethesda). 2024. PMID: 38546739 Free PMC article.
-
Comparative Genomic Analyses of Colletotrichum lindemuthianum Pathotypes with Different Virulence Levels and Lifestyles.J Fungi (Basel). 2024 Sep 13;10(9):651. doi: 10.3390/jof10090651. J Fungi (Basel). 2024. PMID: 39330411 Free PMC article.
-
Genome-wide analysis of Claviceps paspali: insights into the secretome of the main species causing ergot disease in Paspalum spp.BMC Genomics. 2021 Oct 26;22(1):766. doi: 10.1186/s12864-021-08077-0. BMC Genomics. 2021. PMID: 34702162 Free PMC article.
-
Comparative genome analysis indicates high evolutionary potential of pathogenicity genes in Colletotrichum tanaceti.PLoS One. 2019 May 31;14(5):e0212248. doi: 10.1371/journal.pone.0212248. eCollection 2019. PLoS One. 2019. PMID: 31150449 Free PMC article.
References
-
- Bowen N. J., Jordan I. K. (2002). Transposable elements and eukaryotic complexity 65 transposable elements and the evolution of eukaryotic complexity. Curr. Issues Mol. Biol. 4 65–76. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

