STAT2 Signaling Regulates Macrophage Phenotype During Influenza and Bacterial Super-Infection
- PMID: 30337919
- PMCID: PMC6178135
- DOI: 10.3389/fimmu.2018.02151
STAT2 Signaling Regulates Macrophage Phenotype During Influenza and Bacterial Super-Infection
Abstract
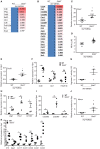
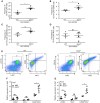
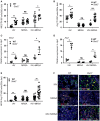
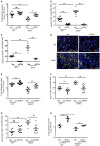
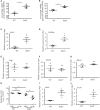
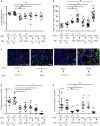
Influenza is a common respiratory virus that infects between 5 and 20% of the US population and results in 30,000 deaths annually. A primary cause of influenza-associated death is secondary bacterial pneumonia. We have previously shown that influenza induces type I interferon (IFN)-mediated inhibition of Type 17 immune responses, resulting in exacerbation of bacterial burden during influenza and Staphylococcus aureus super-infection. In this study, we investigated the role of STAT2 signaling during influenza and influenza-bacterial super-infection in mice. Influenza-infected STAT2-/- mice had increased morbidity, viral burden, and inflammation when compared to wild-type mice. Despite an exaggerated inflammatory response to influenza infection, we found increased bacterial control and survival in STAT2 deficient mice during influenza-MRSA super-infection compared to controls. Further, we found that increased bacterial clearance during influenza-MRSA super-infection is not due to rescue of Type 17 immunity. Absence of STAT2 was associated with increased accumulation of M1, M2 and M1/M2 co-expressing macrophages during influenza-bacterial super-infection. Neutralization of IFNγ (M1) and/or Arginase 1 (M2) impaired bacterial clearance in Stat2-/- mice during super-infection, demonstrating that pulmonary macrophages expressing a mixed M1/M2 phenotype promote bacterial control during influenza-bacterial super-infection. Together, these results suggest that the STAT2 signaling is involved in suppressing macrophage activation and bacterial control during influenza-bacterial super-infection. Further, these studies reveal novel mechanistic insight into the roles of macrophage subpopulations in pulmonary host defense.
Keywords: STAT2; Staphylococcus aureus; influenza; lung; macrophages; pneumonia; super-infection.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

