White Spot Syndrome Virus-Induced Shrimp miR-315 Attenuates Prophenoloxidase Activation via PPAE3 Gene Suppression
- PMID: 30337920
- PMCID: PMC6178132
- DOI: 10.3389/fimmu.2018.02184
White Spot Syndrome Virus-Induced Shrimp miR-315 Attenuates Prophenoloxidase Activation via PPAE3 Gene Suppression
Abstract
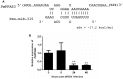
MicroRNAs (miRNAs), the small non-coding RNAs, play a pivotal role in post-transcriptional gene regulation in various cellular processes. However, the miRNA function in shrimp antiviral response is not clearly understood. This research aims to uncover the function of pmo-miR-315, a white spot syndrome virus (WSSV)-responsive miRNAs identified from Penaeus monodon hemocytes during WSSV infection. The expression of the predicted pmo-miR-315 target mRNA, a novel PmPPAE gene called PmPPAE3, was negatively correlated with that of the pmo-miR-315. Furthermore, the luciferase assay indicated that the pmo-miR-315 directly interacted with the target site in PmPPAE3 suggesting the regulatory role of pmo-miR-315 on PmPPAE3 gene expression. Introducing the pmo-miR-315 into the WSSV-infected shrimp caused the reduction of the PmPPAE3 transcript level and, hence, the PO activity activated by the PmPPAE3 whereas the WSSV copy number in the shrimp hemocytes was increased. Taken together, our findings state a crucial role of pmo-miR-315 in attenuating proPO activation via PPAE3 gene suppression and facilitating the WSSV propagation in shrimp WSSV infection.
Keywords: Penaeus monodon; invertebrates; microRNA; prophenoloxidase; viral infection.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

