Meta-Analysis Identification of Highly Robust and Differential Immune-Metabolic Signatures of Systemic Host Response to Acute and Latent Tuberculosis in Children and Adults
- PMID: 30337941
- PMCID: PMC6180280
- DOI: 10.3389/fgene.2018.00457
Meta-Analysis Identification of Highly Robust and Differential Immune-Metabolic Signatures of Systemic Host Response to Acute and Latent Tuberculosis in Children and Adults
Abstract
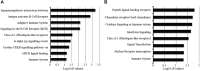
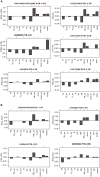
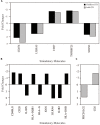
Background: Whole blood expression profiling is a mainstay for delineating differential diagnostic signatures of infection yet is subject to high variability that reduces power and complicates clinical usefulness. To date, confirmatory high confidence expression profiling signatures for clinical use remain uncertain. Here we have sought to evaluate the reproducibility and confirmatory nature of differential expression signatures, comprising molecular and cellular pathways, across multiple international clinical observational studies investigating children and adult whole blood transcriptome responses to tuberculosis (TB). Methods and findings: A systematic search and quality control assessment of gene expression repositories for human TB using whole blood resulted in 11 datasets with a total of 1073 patients from Africa, Europe, and South America. A non-parametric estimation of percentage of false prediction was used for meta-analysis of high confidence differential expression analysis. Deconvolution analysis was applied to infer changes in immune cell proportions and enrichment tests applied using pathway database resources. Meta-analysis identified high confidence differentially expressed genes, comprising 372 in adult active-TB versus latent-TB (LTBI), 332 in adult active-TB versus controls (CON), five in LTBI versus CON, and 415 in childhood active-TB versus LTBI. Notably, these confirmatory markers have low representation in published signatures for diagnosing TB. Pathway biology analysis of high confidence gene sets revealed dominant metabolic and innate-immune pathway signatures while suppressed signatures were enriched with adaptive signaling pathways and reduced proportions of T and B cells. Childhood TB showed uniquely strong inflammasome antagonist signature (IL1RN and ILR2), while adult TB patients exhibit a significant preponderance type I and type II IFN markers. Key limitations of the study include the paucity of data on potential confounders. Conclusion: Meta-analysis identified high confidence confirmatory immune-metabolic and cellular expression signatures across studies regardless of the population resource setting, HIV status and circulating endemic pathogens. Notably, previously identified diagnostic signature markers for TB show limited concordance with the confirmatory meta-analysis. Overall, our results support the use of the confirmatory expression signatures for guiding optimized diagnostic, prognostic, and therapeutic monitoring modalities in TB.
Keywords: bioinformatics; immunity; meta-analysis; microarray; systemic responses; tuberculosis.
Figures






References
-
- Abbas A. R., Baldwin D., Ma Y., Ouyang W., Gurney A., Martin F., et al. (2005). Immune response in silico (IRIS): immune-specific genes identified from a compendium of microarray expression data. Genes Immun. 6 319–331. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

