Identification and characterization of long non-coding RNAs in porcine granulosa cells exposed to 2,3,7,8-tetrachlorodibenzo- p-dioxin
- PMID: 30338064
- PMCID: PMC6180664
- DOI: 10.1186/s40104-018-0288-3
Identification and characterization of long non-coding RNAs in porcine granulosa cells exposed to 2,3,7,8-tetrachlorodibenzo- p-dioxin
Abstract
Background: Long non-coding RNAs (lncRNAs) may regulate gene expression in numerous biological processes including cellular response to xenobiotics. The exposure of living organisms to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), a persistent environmental contaminant, results in reproductive defects in many species including pigs. The aims of the study were to identify and characterize lncRNAs in porcine granulosa cells as well as to examine the effects of TCDD on the lncRNA expression profile in the cells.
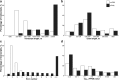
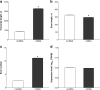
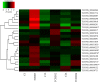
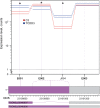
Results: One thousand six hundred sixty-six lncRNAs were identified and characterized in porcine granulosa cells. The identified lncRNAs were found to be shorter than mRNAs. In addition, the number of exons was lower in lncRNAs than in mRNAs and their exons were longer. TCDD affected the expression of 22 lncRNAs (differentially expressed lncRNAs [DELs]; log2 fold change ≥ 1, P-adjusted < 0.05) in the examined cells. Potential functions of DELs were indirectly predicted via searching their target cis- and trans-regulated protein-coding genes. The co-expression analysis revealed that DELs may influence the expression of numerous genes, including those involved in cellular response to xenobiotics, dioxin metabolism, endoplasmic reticulum stress and cell proliferation. Aryl hydrocarbon receptor (AhR) and cytochrome P450 1A1 (CYP1A1) were found among the trans-regulated genes.
Conclusions: These findings indicate that the identified lncRNAs may constitute a part of the regulatory mechanism of TCDD action in granulosa cells. To our knowledge, this is the first study describing lncRNAs in porcine granulosa cells as well as TCDD effects on the lncRNA expression profile. These results may trigger new research directions leading to better understanding of molecular processes induced by xenobiotics in the ovary.
Keywords: AVG-16 cell line; Granulosa cells; Pig; RNA-Seq; TCDD; lncRNAs.
Conflict of interest statement
Not applicable.Not applicable.The authors declare that they have no competing interests.
Figures


References
LinkOut - more resources
Full Text Sources

