Combined study on clastogenic, aneugenic and apoptotic properties of doxorubicin in human cells in vitro
- PMID: 30338246
- PMCID: PMC6180587
- DOI: 10.1186/s40709-018-0089-z
Combined study on clastogenic, aneugenic and apoptotic properties of doxorubicin in human cells in vitro
Abstract
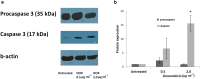
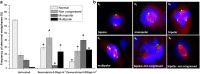
Background: Doxorubicin is a widely used anticancer drug due to its broad spectrum of antitumor activity. Various mechanisms have been proposed for its cytostatic activity, including DNA intercalation, topoisomerase II inhibition, generation of free radicals and apoptosis. The present study aims to further clarify the cytostatic activity of doxorubicin by its specific effect on (a) DNA damage, (b) micronucleation and (c) apoptosis, using a combination of different methods and cell systems such as human lymphocytes and HL-60 human leukemic cells. DNA lesions were analyzed by the alkaline comet assay in combination with formamidopyrimidine (Fpg) and human 8-oxoguanine (hOGG1) repair enzymes. Micronucleation was investigated by the Cytokinesis-Block Micronucleus assay (CBMN) in combination with Fluorescence In Situ Hybridization analysis. Impairment on mitotic apparatus was investigated by double immunofluorescence of β- and γ-tubulin. Apoptotic cell frequency was determined by the CBMN cytome assay. Complementary to the above, caspase-3 level was investigated by Western blot.
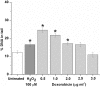
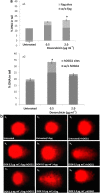
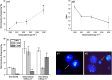
Results: It was found that doxorubicin generates DNA breakage induced by oxidative damage in DNA bases, which can be repaired by the Fpg and hOGG1 enzymes. Increased micronucleus frequency was identified mainly through chromosome breakage and, at a lesser extent, through chromosome delay. Analysis of mitotic spindle showed disturbance of chromosome orientation and centrosome duplication and/or separation, leading to aneuploidy. Enhanced frequency of apoptotic leukemic cells was also observed. Caspase-3 seems to be involved in the generation of apoptosis.
Conclusions: The aforementioned findings derived from different treatment schedules, doses and time of exposure on primary versus transformed cells extend our knowledge about doxorubicin genotoxicity and contribute to the better understanding of the mechanisms by which doxorubicin induces genotoxic effects on human cells.
Keywords: Apoptosis-caspase-3; Doxorubicin; Fpg and hOGG1 comet assay; Micronucleation; Mitotic spindle disturbance.
Figures
Similar articles
-
Aneugenic potential of the nitrogen mustard analogues melphalan, chlorambucil and p-N,N-bis(2-chloroethyl)aminophenylacetic acid in cell cultures in vitro.Mutat Res. 2007 Apr 1;617(1-2):125-37. doi: 10.1016/j.mrfmmm.2007.01.009. Epub 2007 Jan 30. Mutat Res. 2007. PMID: 17324445
-
Comparative study of genetic activity of chlorambucil's active metabolite steroidal esters: the role of steroidal skeleton on aneugenic potential.Mutat Res. 2010 Jul 7;689(1-2):1-11. doi: 10.1016/j.mrfmmm.2010.04.001. Epub 2010 Apr 18. Mutat Res. 2010. PMID: 20403366
-
Okadaic acid: chromosomal non-disjunction analysis in human lymphocytes and study of aneugenic pathway in CHO-K1 cells.Mutat Res. 2005 Oct 15;578(1-2):53-63. doi: 10.1016/j.mrfmmm.2005.02.011. Mutat Res. 2005. PMID: 15885711
-
The in vitro micronucleus test: a multi-endpoint assay to detect simultaneously mitotic delay, apoptosis, chromosome breakage, chromosome loss and non-disjunction.Mutat Res. 1997 Aug 1;392(1-2):19-30. doi: 10.1016/s0165-1218(97)00042-6. Mutat Res. 1997. PMID: 9269328 Review.
-
Inclusion of micronuclei in non-divided mononuclear lymphocytes and necrosis/apoptosis may provide a more comprehensive cytokinesis block micronucleus assay for biomonitoring purposes.Mutagenesis. 2001 Jan;16(1):51-8. doi: 10.1093/mutage/16.1.51. Mutagenesis. 2001. PMID: 11139598 Review.
Cited by
-
PARP Inhibitor Decreases Akt Phosphorylation and Induces Centrosome Amplification and Chromosomal Aneuploidy in CHO-K1 Cells.Int J Mol Sci. 2022 Mar 23;23(7):3484. doi: 10.3390/ijms23073484. Int J Mol Sci. 2022. PMID: 35408845 Free PMC article.
-
Clastogenicity and Aneugenicity of 1,4-Benzoquinone in Different Lineages of Mouse Hematopoietic Stem/Progenitor Cells.Toxics. 2021 May 12;9(5):107. doi: 10.3390/toxics9050107. Toxics. 2021. PMID: 34065823 Free PMC article.
-
Doxorubicin-Induced Translocation of mtDNA into the Nuclear Genome of Human Lymphocytes Detected Using a Molecular-Cytogenetic Approach.Int J Mol Sci. 2020 Oct 17;21(20):7690. doi: 10.3390/ijms21207690. Int J Mol Sci. 2020. PMID: 33080837 Free PMC article.
-
MicroRNA in the Diagnosis and Treatment of Doxorubicin-Induced Cardiotoxicity.Biomolecules. 2023 Mar 20;13(3):568. doi: 10.3390/biom13030568. Biomolecules. 2023. PMID: 36979503 Free PMC article. Review.
-
Encapsulation of Nedaplatin in Novel PEGylated Liposomes Increases Its Cytotoxicity and Genotoxicity against A549 and U2OS Human Cancer Cells.Pharmaceutics. 2020 Sep 10;12(9):863. doi: 10.3390/pharmaceutics12090863. Pharmaceutics. 2020. PMID: 32927897 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials

