Identification of Incidental Germline Mutations in Patients With Advanced Solid Tumors Who Underwent Cell-Free Circulating Tumor DNA Sequencing
- PMID: 30339520
- PMCID: PMC6286162
- DOI: 10.1200/JCO.18.00328
Identification of Incidental Germline Mutations in Patients With Advanced Solid Tumors Who Underwent Cell-Free Circulating Tumor DNA Sequencing
Abstract
Purpose: To determine the potential for detection of incidental germline cancer predisposition mutations through cell-free DNA (cfDNA) analyses in patients who underwent solid tumor somatic mutation evaluation.
Patients and methods: Data were evaluated from 10,888 unselected patients with advanced (stage III/IV) cancer who underwent Guardant360 testing between November 2015 and December 2016. The main outcome was prevalence of putative germline mutations identified among 16 actionable hereditary cancer predisposition genes.
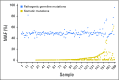
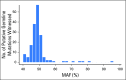
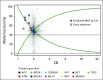
Results: More than 50 cancer types were studied, including lung (41%), breast (19%), colorectal (8%), prostate (6%), pancreatic (3%), and ovarian (2%). Average patient age was 63.5 years (range, 18 to 95 years); 43% were male. One hundred and fifty-six individuals (1.4%) had suspected hereditary cancer mutations in 11 genes. Putative germline mutations were more frequent in individuals younger than 50 years versus those 50 years and older (3.0% v 1.2%, respectively; P < .001). Highest yields of putative germline findings were in patients with ovarian (8.13%), prostate (3.46%), pancreatic (3.34%), and breast (2.2%) cancer. Putative germline mutation identification was consistent among 12 individuals with multiple samples. Patients with circulating tumor DNA copy number variation and/or reversion mutations suggestive of functional loss of the wild-type allele in the tumor DNA also are described.
Conclusion: Detection of putative germline mutations from cfDNA is feasible across multiple genes and cancer types without prior mutation knowledge. Many mutations were found in cancers without clear guidelines for hereditary cancer genetic counseling/testing. Given the clinical significance of identifying hereditary cancer predisposition for patients and their families as well as targetable germline alterations such as in BRCA1 or BRCA2, research on the best way to validate and return potential germline results from cfDNA analysis to clinicians and patients is needed.
Figures

References
-
- National Comprehensive Cancer Network: NCCN clinical practice guidelines in oncology V.1.2017: Genetic/familial high-risk assessment: Colorectal. https://www.nccn.org/professionals/physician_gls/default.aspx#genetics_c....
-
- National Comprehensive Cancer Network: NCCN clinical practice guidelines in oncology V.2.2017: Genetic/familial high-risk assessment: Breast and ovarian. https://www.nccn.org/professionals/physician_gls/default.aspx#genetics_c.... - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

