Cilia-Associated Oxysterols Activate Smoothened
- PMID: 30340023
- PMCID: PMC6503851
- DOI: 10.1016/j.molcel.2018.08.034
Cilia-Associated Oxysterols Activate Smoothened
Abstract
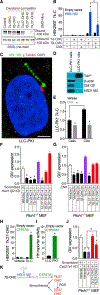
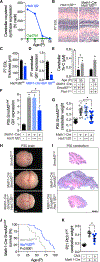
Primary cilia are required for Smoothened to transduce vertebrate Hedgehog signals, but how Smoothened accumulates in cilia and is activated is incompletely understood. Here, we identify cilia-associated oxysterols that promote Smoothened accumulation in cilia and activate the Hedgehog pathway. Our data reveal that cilia-associated oxysterols bind to two distinct Smoothened domains to modulate Smoothened accumulation in cilia and tune the intensity of Hedgehog pathway activation. We find that the oxysterol synthase HSD11β2 participates in the production of Smoothened-activating oxysterols and promotes Hedgehog pathway activity. Inhibiting oxysterol biosynthesis impedes oncogenic Hedgehog pathway activation and attenuates the growth of Hedgehog pathway-associated medulloblastoma, suggesting that targeted inhibition of Smoothened-activating oxysterol production may be therapeutically useful for patients with Hedgehog-associated cancers.
Keywords: CBP; HSD11β2; Hedgehog; Smoothened; cilia; lipid; medulloblastoma; oxysterol; primary cilium; sterol.
Published by Elsevier Inc.
Conflict of interest statement
Declaration of Interests
The authors declare no competing interests.
Figures


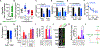
References
-
- Briscoe J, and Therond PP (2013). The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol 14, 416–429. - PubMed
-
- Chavez M, Ena S, Van Sande J, de Kerchove d’Exaerde A, Schurmans, tent regulates trafficking and Sonic Hedgehog signaling output. Dev. Cell 34, 338–350. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 HL020948/HL/NHLBI NIH HHS/United States
- C20724/CRUK_/Cancer Research UK/United Kingdom
- HHMI/Howard Hughes Medical Institute/United States
- R01 GM089740/GM/NIGMS NIH HHS/United States
- 14414/CRUK_/Cancer Research UK/United Kingdom
- R01 HD092659/HD/NICHD NIH HHS/United States
- R01 DK106404/DK/NIDDK NIH HHS/United States
- R01 GM102498/GM/NIGMS NIH HHS/United States
- R01 AR054396/AR/NIAMS NIH HHS/United States
- R01 AR065409/AR/NIAMS NIH HHS/United States
- R01 GM095941/GM/NIGMS NIH HHS/United States
- A14414/CRUK_/Cancer Research UK/United Kingdom
- 26752/CRUK_/Cancer Research UK/United Kingdom
- P30 DK098722/DK/NIDDK NIH HHS/United States
- K08 CA212279/CA/NCI NIH HHS/United States
- 647278/ERC_/European Research Council/International
- T32 HL007731/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

