Structure-based redesign of docking domain interactions modulates the product spectrum of a rhabdopeptide-synthesizing NRPS
- PMID: 30341296
- PMCID: PMC6195595
- DOI: 10.1038/s41467-018-06712-1
Structure-based redesign of docking domain interactions modulates the product spectrum of a rhabdopeptide-synthesizing NRPS
Abstract
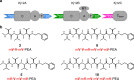
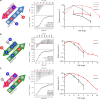
Several peptides in clinical use are derived from non-ribosomal peptide synthetases (NRPS). In these systems multiple NRPS subunits interact with each other in a specific linear order mediated by specific docking domains (DDs), whose structures are not known yet, to synthesize well-defined peptide products. In contrast to classical NRPSs, single-module NRPS subunits responsible for the generation of rhabdopeptide/xenortide-like peptides (RXPs) can act in different order depending on subunit stoichiometry thereby producing peptide libraries. To define the basis for their unusual interaction patterns, we determine the structures of all N-terminal DDs (NDDs) as well as of an NDD-CDD complex and characterize all putative DD interactions thermodynamically for such a system. Key amino acid residues for DD interactions are identified that upon their exchange change the DD affinity and result in predictable changes in peptide production. Recognition rules for DD interactions are identified that also operate in other megasynthase complexes.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
A New Docking Domain Type in the Peptide-Antimicrobial-Xenorhabdus Peptide Producing Nonribosomal Peptide Synthetase from Xenorhabdus bovienii.ACS Chem Biol. 2020 Apr 17;15(4):982-989. doi: 10.1021/acschembio.9b01022. Epub 2020 Mar 25. ACS Chem Biol. 2020. PMID: 32167274
-
Engineering of Specific Single-Module Nonribosomal Peptide Synthetases of the RXP Type for the Production of Defined Peptides.ACS Synth Biol. 2023 Jan 20;12(1):203-212. doi: 10.1021/acssynbio.2c00472. Epub 2022 Dec 19. ACS Synth Biol. 2023. PMID: 36535068 Free PMC article.
-
NMR resonance assignments for a docking domain pair with an attached thiolation domain from the PAX peptide-producing NRPS from Xenorhabdus cabanillasii.Biomol NMR Assign. 2021 Apr;15(1):229-234. doi: 10.1007/s12104-021-10010-1. Epub 2021 Mar 5. Biomol NMR Assign. 2021. PMID: 33675014 Free PMC article.
-
Ways of assembling complex natural products on modular nonribosomal peptide synthetases.Chembiochem. 2002 Jun 3;3(6):490-504. doi: 10.1002/1439-7633(20020603)3:6<490::AID-CBIC490>3.0.CO;2-N. Chembiochem. 2002. PMID: 12325005 Review.
-
[Advances in the study of the mechanism and application of nonribosomal peptide synthetases].Wei Sheng Wu Xue Bao. 2007 Aug;47(4):734-7. Wei Sheng Wu Xue Bao. 2007. PMID: 17944384 Review. Chinese.
Cited by
-
Microbial Lipopeptide-Producing Strains and Their Metabolic Roles under Anaerobic Conditions.Microorganisms. 2021 Sep 25;9(10):2030. doi: 10.3390/microorganisms9102030. Microorganisms. 2021. PMID: 34683351 Free PMC article. Review.
-
Xenorhabdus spp.: An Overview of the Useful Facets of Mutualistic Bacteria of Entomopathogenic Nematodes.Life (Basel). 2022 Aug 31;12(9):1360. doi: 10.3390/life12091360. Life (Basel). 2022. PMID: 36143397 Free PMC article. Review.
-
Biosynthesis of depsipeptides, or Depsi: The peptides with varied generations.Protein Sci. 2020 Dec;29(12):2316-2347. doi: 10.1002/pro.3979. Epub 2020 Nov 2. Protein Sci. 2020. PMID: 33073901 Free PMC article. Review.
-
Complex peptide natural products: Biosynthetic principles, challenges and opportunities for pathway engineering.Synth Syst Biotechnol. 2022 Feb 9;7(1):631-647. doi: 10.1016/j.synbio.2022.01.007. eCollection 2022 Mar. Synth Syst Biotechnol. 2022. PMID: 35224231 Free PMC article.
-
Nostoc edaphicum CCNP1411 from the Baltic Sea-A New Producer of Nostocyclopeptides.Mar Drugs. 2020 Aug 26;18(9):442. doi: 10.3390/md18090442. Mar Drugs. 2020. PMID: 32858999 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

