Porcine circovirus 2 (PCV-2) genetic variability under natural infection scenario reveals a complex network of viral quasispecies
- PMID: 30341330
- PMCID: PMC6195574
- DOI: 10.1038/s41598-018-33849-2
Porcine circovirus 2 (PCV-2) genetic variability under natural infection scenario reveals a complex network of viral quasispecies
Abstract
Porcine circovirus 2 (PCV-2) is a virus characterized by a high evolutionary rate, promoting the potential emergence of different genotypes and strains. Despite the likely relevance in the emergence of new PCV-2 variants, the subtle evolutionary patterns of PCV-2 at the individual-host level or over short transmission chains are still largely unknown. This study aimed to analyze the within-host genetic variability of PCV-2 subpopulations to unravel the forces driving PCV-2 evolution. A longitudinal weekly sampling was conducted on individual animals located in three farms after the first PCV-2 detection. The analysis of polymorphisms evaluated throughout the full PCV-2 genome demonstrated the presence of several single nucleotide polymorphisms (SNPs) especially in the genome region encoding for the capsid gene. The global haplotype reconstruction allowed inferring the virus transmission network over time, suggesting a relevant within-farm circulation. Evidences of co-infection and recombination involving multiple PCV-2 genotypes were found after mixing with pigs originating from other sources. The present study demonstrates the remarkable within-host genetic variability of PCV-2 quasispecies, suggesting the role of the natural selection induced by the host immune response in driving PCV-2 evolution. Moreover, the effect of pig management in multiple genotype coinfections occurrence and recombination likelihood was demonstrated.
Conflict of interest statement
The authors declare no competing interests.
Figures

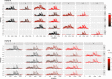
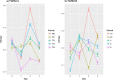
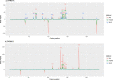
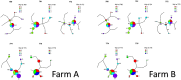

Similar articles
-
Molecular study of Porcine Circovirus type 2 in wild boars and domestic pigs in Uruguay from 2010 to 2014: Predominance of recombinant circulating strains.Gene. 2017 Dec 30;637:230-238. doi: 10.1016/j.gene.2017.09.058. Epub 2017 Sep 28. Gene. 2017. PMID: 28964894
-
Genetic diversity of porcine circovirus type 2 in China between 1999-2017.Transbound Emerg Dis. 2019 Jan;66(1):599-605. doi: 10.1111/tbed.13040. Epub 2018 Nov 1. Transbound Emerg Dis. 2019. PMID: 30307710
-
Genetic typing of porcine circovirus type 2 (PCV-2) isolates from Slovakia.Res Vet Sci. 2011 Feb;90(1):168-73. doi: 10.1016/j.rvsc.2010.04.022. Epub 2010 May 23. Res Vet Sci. 2011. PMID: 20580978
-
The natural history of porcine circovirus type 2: from an inoffensive virus to a devastating swine disease?Vet Microbiol. 2013 Jul 26;165(1-2):13-20. doi: 10.1016/j.vetmic.2012.12.033. Epub 2013 Jan 17. Vet Microbiol. 2013. PMID: 23380460 Review.
-
Porcine Circovirus Type 2 (PCV2) Vaccines in the Context of Current Molecular Epidemiology.Viruses. 2017 May 6;9(5):99. doi: 10.3390/v9050099. Viruses. 2017. PMID: 28481275 Free PMC article. Review.
Cited by
-
Structural characterization of the PCV2d virus-like particle at 3.3 Å resolution reveals differences to PCV2a and PCV2b capsids, a tetranucleotide, and an N-terminus near the icosahedral 3-fold axes.Virology. 2019 Nov;537:186-197. doi: 10.1016/j.virol.2019.09.001. Epub 2019 Sep 3. Virology. 2019. PMID: 31505320 Free PMC article.
-
Genetic Diversity of Porcine Circovirus 2 in Wild Boar and Domestic Pigs in Ukraine.Viruses. 2022 Apr 28;14(5):924. doi: 10.3390/v14050924. Viruses. 2022. PMID: 35632666 Free PMC article.
-
Efficacy Studies of a Trivalent Vaccine Containing PCV-2a, PCV-2b Genotypes and Mycoplasma hyopneumoniae When Administered at 3 Days of Age and 3 Weeks Later against Porcine Circovirus 2 (PCV-2) Infection.Vaccines (Basel). 2022 Aug 1;10(8):1234. doi: 10.3390/vaccines10081234. Vaccines (Basel). 2022. PMID: 36016122 Free PMC article.
-
Porcine circovirus 2 (PCV-2) genotype update and proposal of a new genotyping methodology.PLoS One. 2018 Dec 6;13(12):e0208585. doi: 10.1371/journal.pone.0208585. eCollection 2018. PLoS One. 2018. PMID: 30521609 Free PMC article.
-
Porcine circovirus 3: a new challenge to explore.Front Vet Sci. 2024 Apr 24;10:1266499. doi: 10.3389/fvets.2023.1266499. eCollection 2023. Front Vet Sci. 2024. PMID: 38720992 Free PMC article. Review.
References
-
- Juhan NM, LeRoith T, Opriessnig T, Meng XJ. The open reading frame 3 (ORF3) of porcine circovirus type 2 (PCV2) is dispensable for virus infection but evidence of reduced pathogenicity is limited in pigs infected by an ORF3-null PCV2 mutant. Virus Res. 2010;147:60–66. doi: 10.1016/j.virusres.2009.10.007. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

