TRIP13 and APC15 drive mitotic exit by turnover of interphase- and unattached kinetochore-produced MCC
- PMID: 30341343
- PMCID: PMC6195577
- DOI: 10.1038/s41467-018-06774-1
TRIP13 and APC15 drive mitotic exit by turnover of interphase- and unattached kinetochore-produced MCC
Abstract
The mitotic checkpoint ensures accurate chromosome segregation through assembly of the mitotic checkpoint complex (MCC), a soluble inhibitor of the anaphase-promoting complex/cyclosome (APC/C) produced by unattached kinetochores. MCC is also assembled during interphase by Mad1/Mad2 bound at nuclear pores, thereby preventing premature mitotic exit prior to kinetochore maturation and checkpoint activation. Using degron tagging to rapidly deplete the AAA+ ATPase TRIP13, we show that its catalytic activity is required to maintain a pool of open-state Mad2 for MCC assembly, thereby supporting mitotic checkpoint activation, but is also required for timely mitotic exit through catalytic disassembly of MCC. Strikingly, combining TRIP13 depletion with elimination of APC15-dependent Cdc20 ubiquitination/degradation results in a complete inability to exit mitosis, even when MCC assembly at unattached kinetochores is prevented. Thus, mitotic exit requires MCC produced either in interphase or mitosis to be disassembled by TRIP13-catalyzed removal of Mad2 or APC15-driven ubiquitination/degradation of its Cdc20 subunit.
Conflict of interest statement
The authors declare no competing interests.
Figures

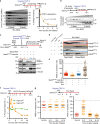
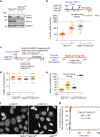

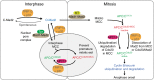
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM029513/GM/NIGMS NIH HHS/United States
- K99 CA218871/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)/International
- K99 CA218871/CA/NCI NIH HHS/United States
- R35 GM122476/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)/International
- R01 GM104141/GM/NIGMS NIH HHS/United States
- R01 GM104141/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)/International
- R01 GM2951/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)/International
- R35 GM122476/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

