Crystal structure of AdoMet radical enzyme 7-carboxy-7-deazaguanine synthase from Escherichia coli suggests how modifications near [4Fe-4S] cluster engender flavodoxin specificity
- PMID: 30341796
- PMCID: PMC6295903
- DOI: 10.1002/pro.3529
Crystal structure of AdoMet radical enzyme 7-carboxy-7-deazaguanine synthase from Escherichia coli suggests how modifications near [4Fe-4S] cluster engender flavodoxin specificity
Abstract
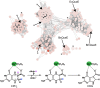
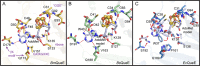
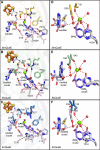
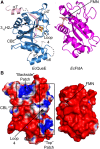
7-Carboxy-7-deazaguanine synthase, QueE, catalyzes the radical mediated ring contraction of 6-carboxy-5,6,7,8-tetrahydropterin, forming the characteristic pyrrolopyrimidine core of all 7-deazaguanine natural products. QueE is a member of the S-adenosyl-L-methionine (AdoMet) radical enzyme superfamily, which harnesses the reactivity of radical intermediates to perform challenging chemical reactions. Members of the AdoMet radical enzyme superfamily utilize a canonical binding motif, a CX3 CXϕC motif, to bind a [4Fe-4S] cluster, and a partial (β/α)6 TIM barrel fold for the arrangement of AdoMet and substrates for catalysis. Although variations to both the cluster-binding motif and the core fold have been observed, visualization of drastic variations in the structure of QueE from Burkholderia multivorans called into question whether a re-haul of the defining characteristics of this superfamily was in order. Surprisingly, the structure of QueE from Bacillus subtilis revealed an architecture more reminiscent of the classical AdoMet radical enzyme. With these two QueE structures revealing varying degrees of alterations to the classical AdoMet fold, a new question arises: what is the purpose of these alterations? Here, we present the structure of a third QueE enzyme from Escherichia coli, which establishes the middle range of the spectrum of variation observed in these homologs. With these three homologs, we compare and contrast the structural architecture and make hypotheses about the role of these structural variations in binding and recognizing the biological reductant, flavodoxin. Broader impact statement: We know more about how enzymes are tailored for catalytic activity than about how enzymes are tailored to react with a physiological reductant. Here, we consider structural differences between three 7-carboxy-7-deazaguanine synthases and how these differences may be related to the interaction between these enzymes and their biological reductant, flavodoxin.
Keywords: AdoMet radical enzymes; flavin mononucleotide; flavodoxin; iron-sulfur clusters; physiological reductant.
© 2018 The Protein Society.
Figures





Similar articles
-
Radical SAM enzyme QueE defines a new minimal core fold and metal-dependent mechanism.Nat Chem Biol. 2014 Feb;10(2):106-12. doi: 10.1038/nchembio.1426. Epub 2013 Dec 22. Nat Chem Biol. 2014. PMID: 24362703 Free PMC article.
-
Radical S-adenosylmethionine enzyme coproporphyrinogen III oxidase HemN: functional features of the [4Fe-4S] cluster and the two bound S-adenosyl-L-methionines.J Biol Chem. 2005 Aug 12;280(32):29038-46. doi: 10.1074/jbc.M501275200. Epub 2005 Jun 20. J Biol Chem. 2005. PMID: 15967800
-
Spectroscopic, steady-state kinetic, and mechanistic characterization of the radical SAM enzyme QueE, which catalyzes a complex cyclization reaction in the biosynthesis of 7-deazapurines.Biochemistry. 2013 Jan 8;52(1):188-98. doi: 10.1021/bi301156w. Epub 2012 Dec 24. Biochemistry. 2013. PMID: 23194065 Free PMC article.
-
Structural diversity in the AdoMet radical enzyme superfamily.Biochim Biophys Acta. 2012 Nov;1824(11):1178-95. doi: 10.1016/j.bbapap.2012.04.006. Epub 2012 Apr 28. Biochim Biophys Acta. 2012. PMID: 22579873 Free PMC article. Review.
-
The novel structure and chemistry of iron-sulfur clusters in the adenosylmethionine-dependent radical enzyme biotin synthase.Arch Biochem Biophys. 2005 Jan 1;433(1):312-21. doi: 10.1016/j.abb.2004.10.003. Arch Biochem Biophys. 2005. PMID: 15581586 Review.
Cited by
-
Not all 5'-deoxyadenosines are created equal: Tracing the provenance of 5'-deoxyadenosine formed by the radical S-adenosyl-L-methionine enzyme 7-carboxy-7-deazaguanine synthase.J Biol Chem. 2025 Apr;301(4):108347. doi: 10.1016/j.jbc.2025.108347. Epub 2025 Feb 25. J Biol Chem. 2025. PMID: 40015645 Free PMC article.
-
Analysis of Electrochemical Properties of S-Adenosyl-l-methionine and Implications for Its Role in Radical SAM Enzymes.J Am Chem Soc. 2019 Jul 17;141(28):11019-11026. doi: 10.1021/jacs.9b00933. Epub 2019 Jul 8. J Am Chem Soc. 2019. PMID: 31283208 Free PMC article.
-
Eukaryotic TYW1 Is a Radical SAM Flavoenzyme.Biochemistry. 2021 Jul 13;60(27):2179-2185. doi: 10.1021/acs.biochem.1c00349. Epub 2021 Jun 29. Biochemistry. 2021. PMID: 34184886 Free PMC article.
-
Journey on the Radical SAM Road as an Accidental Pilgrim.ACS Bio Med Chem Au. 2022 Jun 15;2(3):187-195. doi: 10.1021/acsbiomedchemau.1c00059. Epub 2022 Feb 28. ACS Bio Med Chem Au. 2022. PMID: 35726327 Free PMC article. Review.
-
Evidence for Porphyrin-Mediated Electron Transfer in the Radical SAM Enzyme HutW.Biochemistry. 2023 Mar 21;62(6):1191-1196. doi: 10.1021/acs.biochem.2c00474. Epub 2023 Mar 6. Biochemistry. 2023. PMID: 36877586 Free PMC article.
References
-
- Fujii K, Huennekens F (1974) Activation of methionine synthetase by a reduced triphosphopyridine nucleotide‐dependent flavoprotein system. J Biol Chem 249:6745–6753. - PubMed
-
- Knappe J, Schacht J, Möckel W, Höpner T, Vetter H Jr, Edenharder R (1969) Pyruvate formate‐lyase reaction in Escherichia coli: The enzymatic system converting and inactive form of the lyase into the catalytically active enzyme. Eur J Biochem 11:316–327. - PubMed
-
- Jarrett JT (2003) The generation of 5′‐deoxyadenosyl radicals by adenosylmethionine‐dependent radical enzymes. Curr Opin Chem Biol 7:174–182. - PubMed
-
- Harder J, Eliasson R, Pontis E, Ballinger MD, Reichard P (1992) Activation of the anaerobic ribonucleotide reductase from Escherichia coli by S‐Adenosylmethionine. J Biol Chem 267:25548–25552. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

