Dimerization of an aptamer generated from Ligand-guided selection (LIGS) yields a high affinity scaffold against B-cells
- PMID: 30342154
- PMCID: PMC6248343
- DOI: 10.1016/j.bbagen.2018.10.006
Dimerization of an aptamer generated from Ligand-guided selection (LIGS) yields a high affinity scaffold against B-cells
Abstract
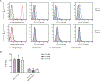
Nucleic Acid Aptamers (NAAs) are a class of synthetic DNA or RNA molecules that bind specifically to their target. We recently introduced an aptamer termed R1.2 against membrane Immunoglobulin M (mIgM) expressing B-cell neoplasms using Ligand Guided Selection (LIGS). While LIGS-generated aptamers are highly specific, their lower affinity prevents aptamers from being used for translational applications. Highly specific aptamers with higher affinity can increase targetability, boosting the application of aptamers as diagnostic and therapeutic molecules. Herein, we report that dimerization of R1.2, an aptamer generated from LIGS, leads to high affinity variants without compromising the specificity. Three dimeric aptamer analogues with variable linker lengths were designed to evaluate the effect of linker length in affinity. The optimized dimeric R1.2 against cultured B-cell neoplasms, four donor B-cell samples and mIgM-positive Waldenström's Macroglobulinemia (WM) showed specificity. Furthermore, confocal imaging of dimeric aptamer and anti-IgM antibody in purified B-cells suggests co-localization. Binding assays against IgM knockout Burkitt's Lymphoma cells utilizing CRISPR/Cas9 further validated specificity of dimeric R1.2. Collectively, our findings show that LIGS-generated aptamers can be re-engineered into dimeric aptamers with high specificity and affinity, demonstrating wide-range of applicability of LIGS in developing clinically practical diagnostic and therapeutic aptamers.
Keywords: Aptamer; B-cell lymphoma; Dimerization; Ligand-guided selection; mIgM.
Copyright © 2018 Elsevier B.V. All rights reserved.
Conflict of interest statement
Disclosures/Conflict of Interest
Authors declare no competing interests
Figures



References
-
- Harris NL, Jaffe ES, Stein H, Banks PM, Chan JK, Cleary ML, Delsol G, De Wolf-Peeters C, Falini B, Gatter KC, et al. , A revised European-American classification of lymphoid neoplasms: a proposal from the International Lymphoma Study Group, Blood, 84 (1994) 1361–1392. - PubMed
-
- Perry AM, Diebold J, Nathwani BN, MacLennan KA, Muller-Hermelink HK, Bast M, Boilesen E, Armitage JO, Weisenburger DD, Non-Hodgkin lymphoma in the developing world: review of 4539 cases from the International Non-Hodgkin Lymphoma Classification Project, Haematologica, 101 (2016) 1244–1250. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

