Revisiting Dscam diversity: lessons from clustered protocadherins
- PMID: 30343321
- PMCID: PMC11105660
- DOI: 10.1007/s00018-018-2951-4
Revisiting Dscam diversity: lessons from clustered protocadherins
Abstract
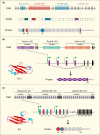
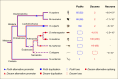
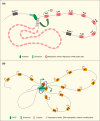
The complexity of neuronal wiring relies on the extraordinary recognition diversity of cell surface molecules. Drosophila Dscam1 and vertebrate clustered protocadherins (Pcdhs) are two classic examples of the striking diversity from a complex genomic locus, wherein the former encodes more than 10,000 distinct isoforms via alternative splicing, while the latter employs alternative promoters to attain isoform diversity. These structurally unrelated families show remarkably striking molecular parallels and even similar functions. Recent studies revealed a novel Dscam gene family with tandemly arrayed 5' cassettes in Chelicerata (e.g., the scorpion Mesobuthus martensii and the tick Ixodes scapularis), similar to vertebrate clustered Pcdhs. Likewise, octopus shows a more remarkable expansion of the Pcdh isoform repertoire than human. These discoveries of Dscam and Pcdh diversification reshape the evolutionary landscape of recognition molecule diversity and provide a greater understanding of convergent molecular strategies for isoform diversity. This article reviews new insights into the evolution, regulatory mechanisms, and functions of Dscam and Pcdh isoform diversity. In particular, the convergence of clustered Dscams and Pcdhs is highlighted.
Keywords: Down syndrome cell adhesion molecule; Duplication; Homophilic binding; Neural circuit; Pcdh; Regulatory mechanism.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

