Deamidation of Amino Acids on the Surface of Adeno-Associated Virus Capsids Leads to Charge Heterogeneity and Altered Vector Function
- PMID: 30343890
- PMCID: PMC6277538
- DOI: 10.1016/j.ymthe.2018.09.013
Deamidation of Amino Acids on the Surface of Adeno-Associated Virus Capsids Leads to Charge Heterogeneity and Altered Vector Function
Abstract
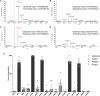
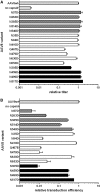
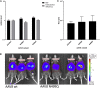
Post-translational modification of the adeno-associated virus capsids is a poorly understood factor in the development of these viral vectors into pharmaceutical products. Here we report the extensive capsid deamidation of adeno-associated virus serotype 8 and seven other diverse adeno-associated virus serotypes, with supporting evidence from structural, biochemical, and mass spectrometry approaches. The extent of deamidation at each site depended on the vector's age and multiple primary-sequence and three-dimensional structural factors. However, the extent of deamidation was largely independent of the vector recovery and purification conditions. We demonstrate the potential for deamidation to impact transduction activity and, moreover, correlate an early time point loss in vector activity to rapidly progressing spontaneous deamidation at several adeno-associated virus 8 asparagines. We explore mutational strategies that stabilize side-chain amides, improving vector transduction and reducing the lot-to-lot molecular variability that presents a key concern in biologics manufacturing. This study illuminates a previously unknown aspect of adeno-associated virus capsid heterogeneity and highlights its importance in the development of these vectors for gene therapy.
Keywords: adeno-associated virus; bioengineering; gene therapy; post-translational modification; structural biology; virus structure.
Copyright © 2018 The American Society of Gene and Cell Therapy. Published by Elsevier Inc. All rights reserved.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

