TCP Transcription Factors in Moso Bamboo (Phyllostachys edulis): Genome-Wide Identification and Expression Analysis
- PMID: 30344527
- PMCID: PMC6182085
- DOI: 10.3389/fpls.2018.01263
TCP Transcription Factors in Moso Bamboo (Phyllostachys edulis): Genome-Wide Identification and Expression Analysis
Abstract
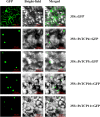
TEOSINTE BRANCHED 1, CYCLOIDEA, and PROLIFERATING CELL FACTORS (T), members of a plant-specific gene family, play significant roles during plant growth and development, as well as in response to environmental stress. However, knowledge about this family in moso bamboo (Phyllostachys edulis) is limited. Therefore, in this study, the first genome-wide identification, classification, characterization, and expression pattern analysis of the TCP transcription factor family in moso bamboo was performed. Sixteen TCP members were identified from the moso bamboo genome using a BLASTP algorithm-based method and verified using the Pfam database. Based on a multiple-sequence alignment, the members were divided into two subfamilies, and members of the same family shared highly conserved motif structures. Subcellular localization and transactivation activity analyses of four selected genes revealed that they were nuclear localized and had self-activation activities. Additionally, the expression levels of several PeTCP members were significantly upregulated under abscisic acid, methyl jasmonate, and salicylic acid treatments, indicating that they play crucial plant hormone transduction roles in the processes of plant growth and development, as well as in responses to environmental stresses. Thus, the current study provides previously lacking information on the TCP family in moso bamboo and reveals the potential functions of this gene family in growth and development.
Keywords: TCP transcription factors; expression patterns; moso bamboo (Phyllostachys edulis); subcellular localization; transcription activity.
Figures





Similar articles
-
Identification of TCP family in moso bamboo (Phyllostachys edulis) and salt tolerance analysis of PheTCP9 in transgenic Arabidopsis.Planta. 2022 Jun 7;256(1):5. doi: 10.1007/s00425-022-03917-z. Planta. 2022. PMID: 35670871
-
Genome-Wide Identification and Expression Analyses of the bZIP Transcription Factor Genes in moso bamboo (Phyllostachys edulis).Int J Mol Sci. 2019 May 5;20(9):2203. doi: 10.3390/ijms20092203. Int J Mol Sci. 2019. PMID: 31060272 Free PMC article.
-
Genome-Wide Identification and Characterization of the UBP Gene Family in Moso Bamboo (Phyllostachys edulis).Int J Mol Sci. 2019 Sep 3;20(17):4309. doi: 10.3390/ijms20174309. Int J Mol Sci. 2019. PMID: 31484390 Free PMC article.
-
Genome-wide identification and expression analysis of LBD transcription factor genes in Moso bamboo (Phyllostachys edulis).BMC Plant Biol. 2021 Jun 28;21(1):296. doi: 10.1186/s12870-021-03078-3. BMC Plant Biol. 2021. PMID: 34182934 Free PMC article.
-
Class I TCP in fruit development: much more than growth.Front Plant Sci. 2024 May 28;15:1411341. doi: 10.3389/fpls.2024.1411341. eCollection 2024. Front Plant Sci. 2024. PMID: 38863555 Free PMC article. Review.
Cited by
-
Genome-Wide Identification, Characterization, and Expression of TCP Genes Family in Orchardgrass.Genes (Basel). 2023 Apr 16;14(4):925. doi: 10.3390/genes14040925. Genes (Basel). 2023. PMID: 37107682 Free PMC article.
-
Genome-wide identification and expression analysis of the TCP transcription factor family and its response to abiotic stress in rapeseed (Brassica napus L.).3 Biotech. 2025 May;15(5):119. doi: 10.1007/s13205-025-04273-x. Epub 2025 Apr 7. 3 Biotech. 2025. PMID: 40201755 Free PMC article.
-
Organelle Visualization With Multicolored Fluorescent Markers in Bamboo.Front Plant Sci. 2021 Apr 15;12:658836. doi: 10.3389/fpls.2021.658836. eCollection 2021. Front Plant Sci. 2021. PMID: 33936145 Free PMC article.
-
TCP Transcription Factors Involved in Shoot Development of Ma Bamboo (Dendrocalamus latiflorus Munro).Front Plant Sci. 2022 May 10;13:884443. doi: 10.3389/fpls.2022.884443. eCollection 2022. Front Plant Sci. 2022. PMID: 35620688 Free PMC article.
-
The G2-Like gene family in Populus trichocarpa: identification, evolution and expression profiles.BMC Genom Data. 2023 Jul 5;24(1):37. doi: 10.1186/s12863-023-01138-1. BMC Genom Data. 2023. PMID: 37403017 Free PMC article.
References
-
- Biłas R., Szafran K., Hnatuszko-Konka K., Kononowicz A. K. (2016). Cis-regulatory elements used to control gene expression in plants. Plant Cell Tissue Organ Cult. 127 1–19. 10.1007/s11240-016-1057-7 - DOI
-
- Broholm S. K., Tähtiharju S., Laitinen R. A. E., Albert V. A., Teeri T. H., Elomaa P. (2008). A TCP domain transcription factor controls flower type specification along the radial axis of the Gerbera (Asteraceae) inflorescence. Proc. Natl. Acad. Sci. U.S.A. 105 9117–9122. 10.1073/pnas.0801359105 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

